Figure 4. Motifs cluster into a low-dimensional set of basis motifs.
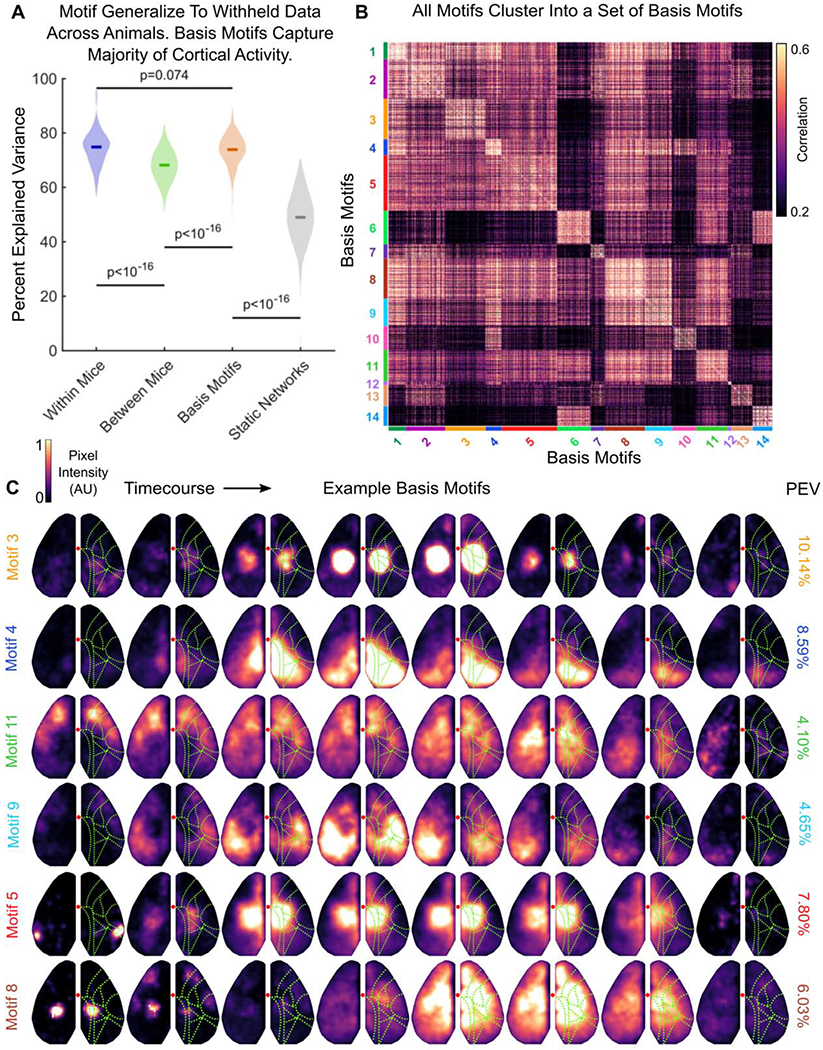
(A) Comparison of the percent of variance in neural activity explained by motifs from the same mouse (within; purple), by motifs from other mice (between; green), by basis motifs (orange), and by static network versions of basis motifs (gray). Static networks for each basis motifs were derived as in Figure 3 (see STAR Methods). All show fit to withheld data (N=144). Full distribution shown; dark lines indicate median. Horizontal lines indicate pairwise comparisons. All p-values estimated with Mann-Whitney U-test.
(B) Pairwise peak cross-correlation between all 2622 discovered motifs. Motifs are grouped by their membership in basis motif clusters. Basis motif identity is indicated with color code along axes. Group numbering (and thus the sorting of the correlation matrix) is determined by relative variance explained by each basis motif.
(C) Representative timecourses for example basis motifs. Example basis motifs were chosen to display the diversity of patterns observed in motifs. See Figure S3 for all basis motifs, Movie S2 for full motif timecourses, and Table S2 for written description of motifs. Display follows Figure 1D, except without direction of flow arrows. Right column shows the relative percent explained variance in neural activity captured by each basis motif.
See also Figures S3, S4, S5, and Tables S1, S2, S3, and Video S2.
