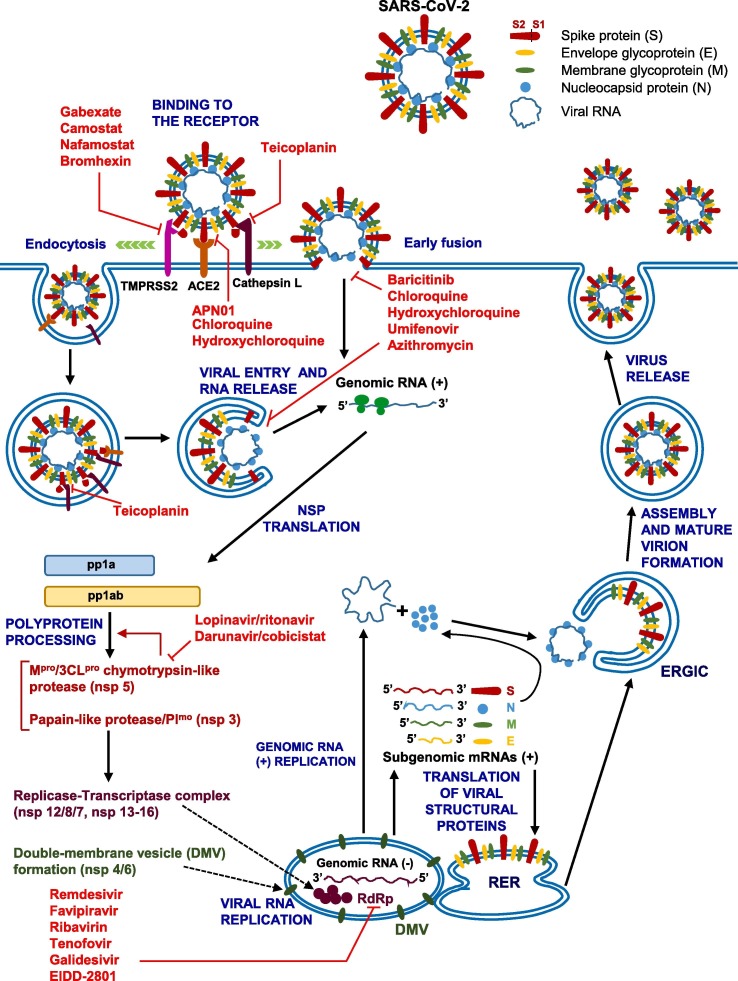
Fig. 1.
Schematic diagram of SARS-CoV-2 replication cycle in human cells and potential viral targets of repurposed drugs that have been empirically used and tested in clinical trials for COVID-19 treatment. During the viral replication cycle, SARS-CoV-2 spike (S) protein binds to ACE2 in host cells and after the attachment step, the entry process requires the S protein priming by cellular proteases (i.e., TMPRSS2, cathepsin L, furin). Fusion of the virus and cell membranes likely occurs both at the plasma membrane (early fusion) and endosomal level (endocytosis) after which the release of the nucleocapsid into the cytoplasm takes place. Most of viral genome sequence is directly translated to produce the polyproteins pp1a and pp1ab, which are processed by viral proteases (3CLpro/Mpro, PLpro) into 16 nonstructural proteins (nsps), including RNA-dependent RNA polymerase (RdRp) and other proteins that form the replication-transcription complex, which is anchored to double-membrane vesicles (DMV) integrated into a reticulovesicular network of modified endoplasmic reticulum membranes. The viral RdRp synthesizes a full-length complementary negative-strand RNA as template for the production of positive-strand genome of the virus progeny and a set of subgenomic mRNAs deriving from negative-sense RNA intermediates (not shown). Subgenomic mRNAs are translated into structural proteins in the rough endoplasmic reticulum (RER) [spike (S), membrane (M), envelope (E) proteins], or in the cytosol [nucleocapsid (N) protein]. The S, E and M move along the intermediate compartment of the endoplasmic reticulum-Golgi (ERGIC). The viral genomic RNA is encapsulated by the nucleocapsid N protein and, thereafter, buds into the ERGIC and acquires a membrane containing the S, E and M structural proteins. Finally, the virus is released by exocytosis. Blunt red arrows indicate the potential targets of the listed drugs. See the text for further details. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)