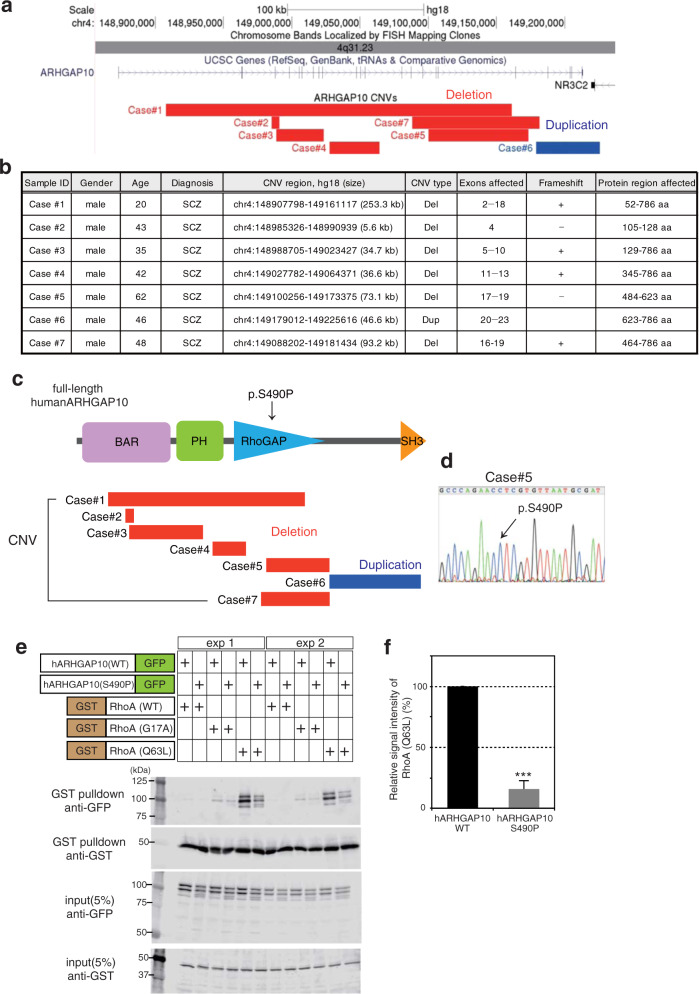
Fig. 1. Exonic CNVs in ARHGAP10 and biological features.
a The exonic CNVs in ARHGAP10 identified in this study. Exonic deletions in ARHGAP10, indicated by the red boxes, were identified in six patients with SCZ. Exonic duplication, indicated by the blue box, was identified in Case#6. b Details of rare exonic CNVs of ARHGAP10 identified in this study. Genomic locations are given in NCBI build 36/UCSC hg18 coordinates. Exons and protein region affected by exonic CNVs are based on NM_024605.3 and NP_078881.3, respectively. c The structure of human ARHGAP10 and the locations of the missense variant (p.S490P) and exonic CNVs identified in patients with SCZ. d Sanger sequencing results of the prioritized variant p.S490P. A missense variant (p.S490P) identified in Case#5. This variant was within the exonic deletion of ARHGAP10 on the other allele and located at the RhoGAP domain. e GST binding assay using GST-RhoA, GST-RhoA (p.G17A), and GST-RhoA (p.Q63L) as bait proteins and GFP-tagged human ARHGAP10 full-length wild-type (WT) and ARHGAP10 (p.S490P) mutant as pray proteins. Input and bound proteins were detected on immunoblots probed with the anti-GST antibody. Densitometric quantification of immunoblots using Odyssey Clx (LI-COR, USA). f Band intensity percentage is presented relative to the corresponding bait (15.8% ± S.E.M. 3.95%, p = 0.0002) (three independent experiments).