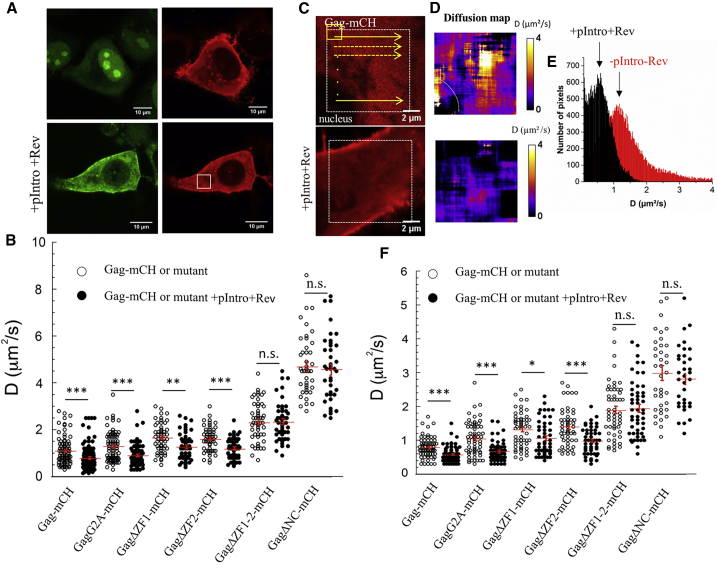
Figure 8.
RICS analysis of Gag-mCH diffusion in the cytoplasm. (A) Shown are confocal images of MS2-eGFP and Gag-mCH expressing cells in the absence (top panels) and the presence (bottom panels) of pIntro and Rev. MS2-eGFP-gRNA was observed in the cell cytoplasm (green channel), and the RICS measurements were performed on the labeled Gag proteins in the red channel. (B) Shown are diffusion coefficient values of Gag proteins in the presence and the absence of pIntro and Rev. (C) Shown are confocal images and (D) corresponding diffusion maps of Gag-mCH in the absence (top panels) and the presence (bottom panels) of pIntro and Rev. (E) Shown is a histogram representation of the D values of the diffusion maps. The arrows show the positions of the most frequent D values, called Dmax. (F) Dmax values of Gag proteins in the presence and absence of pIntro and Rev are shown. In (B) and (D), the measured values, the mean values, and the corresponding SEM of 50–60 measurements in three independent experiments (15–20 cells analyzed per experiment) are indicated. The statistical analysis was realized by a Student’s t-test with significant differences represented by ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. To see this figure in color, go online.