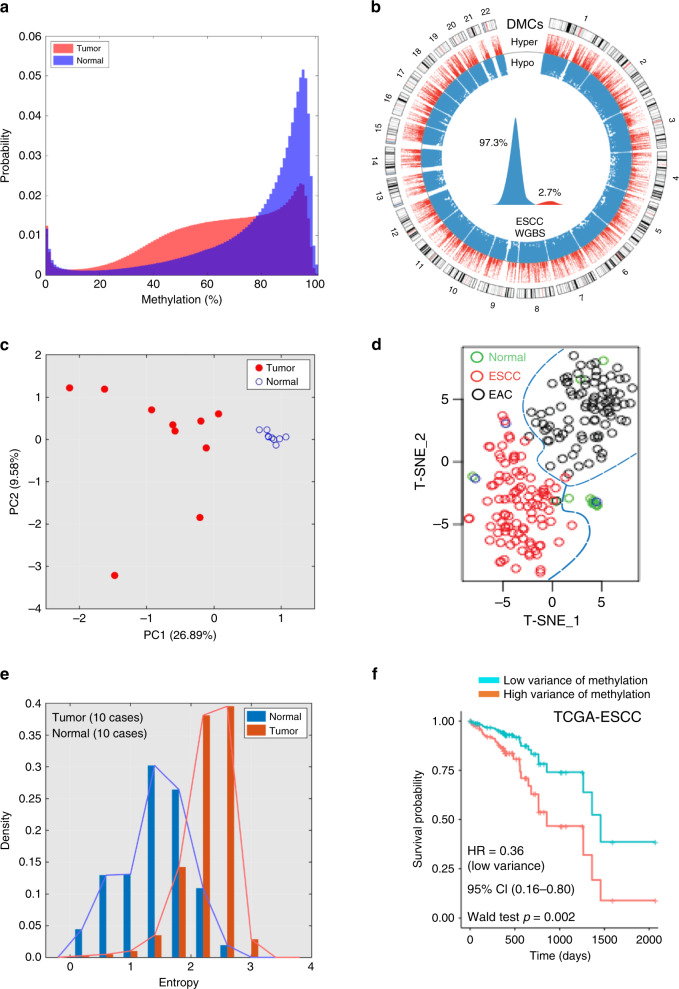
Fig. 1. Epigenetic landscape and heterogeneity in esophageal squamous cell carcinoma (ESCC).
a Ten pairs of ESCC and adjacent normal tissues were performed whole-genome bisulfite sequencing (WGBS). The asymmetric density distribution of all CpG methylation statuses in the normal esophageal tissues versus ESCC. ESCCs lose methylation which leaves most CpGs partially methylated. Normal = blue, tumor = red. b Circos plot of >5 million differentially methylated CpGs (DMCs) between ESCC tumor and adjacent normal tissue. DMCs are substantially hypomethylated in ESCC (97.3%). Only 2.7% are hypermethylated in ESCC. c Principal component analysis (PCA) shows that characteristic CpGs discriminate tumor samples from normal samples. d t-Distributed Stochastic Neighbor Embedding (t-SNE) showed CpG methylation profiling of TCGA-esophageal cancer from human methylation 450K analysis clustered into either normal tissue (n = 15, green circles) or ESCC (n = 97, red circles) or esophageal adenocarcinoma (n = 89, black circles) subtypes. e Entropy analysis of all CpGs showed variations per CpG in normal esophageal tissues (blue bars) and ESCC (red bars). The entropy of CpGs in ESCC was higher than in normal samples. f Multivariate cox proportional hazard analysis demonstrated TCGA-ESCC patients (n = 92) with lower variance of CpG methylation in tumors showed better survival time than those with higher variance. Median variance was used to discriminate high versus low variance groups. Variance of DNA methylation changes were normalized for age, gender, and alcohol consumption in patients. Statistical significance was assessed by two-sided Wald test, p = 0.002.