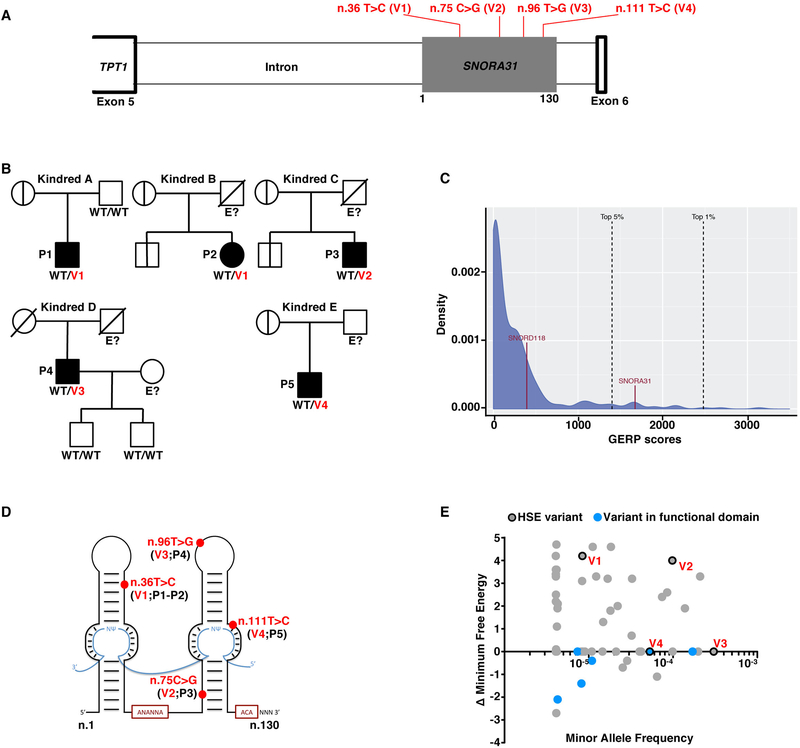
Figure 1. Heterozygous SNORA31 mutations in herpes simplex encephalitis patients from five unrelated kindreds.
A) Schematic representation of the genomic structure of human SNORA31. Human SNORA31 is located on chromosome 13, between exons 5 and 6 of the host gene TPT1. The mutations found in five HSE patients are shown in red: n.36T>C (V1) in patient 1 (P1) and P2; n.75C>G (V2) in P3; n.96T>G (V3) in P4; n.111T>C (V4) in P5. B) Family pedigrees with allele segregation in the five families. The patients, in black, are heterozygous for the following mutations (‘V’ in red): n.36T>C (V1) in kindreds A and B; n.75C>G (V2) in kindred C; n.96T>G (V3) in kindred D; n.111T>C (V4) in kindred E. Vertical bars indicate the same SNORA31 genotype as the patient from the corresponding family. “E?” indicates that the individual’s SNORA31 genotype is unknown. C) Conservation score ranking of the known human snoRNA genes, as assessed by the GERP++ method. Density (y-axis) of GERP scores (x-axis) for conserved elements overlapping snoRNAs. D) Schematic representation of the canonical secondary structure of H/ACA class snoRNAs, including snoRNA31. The positions of the patients’ SNORA31 variants are indicated in red. E) Frequency and predicted impact on the secondary structure of snoRNA31, as measured by the calculated change in minimum free energy of mutant sequences relative to wild type, for all variants found in gnomAD. All variants associated with a change in minimum free energy of more than 1 were considered possibly damaging.