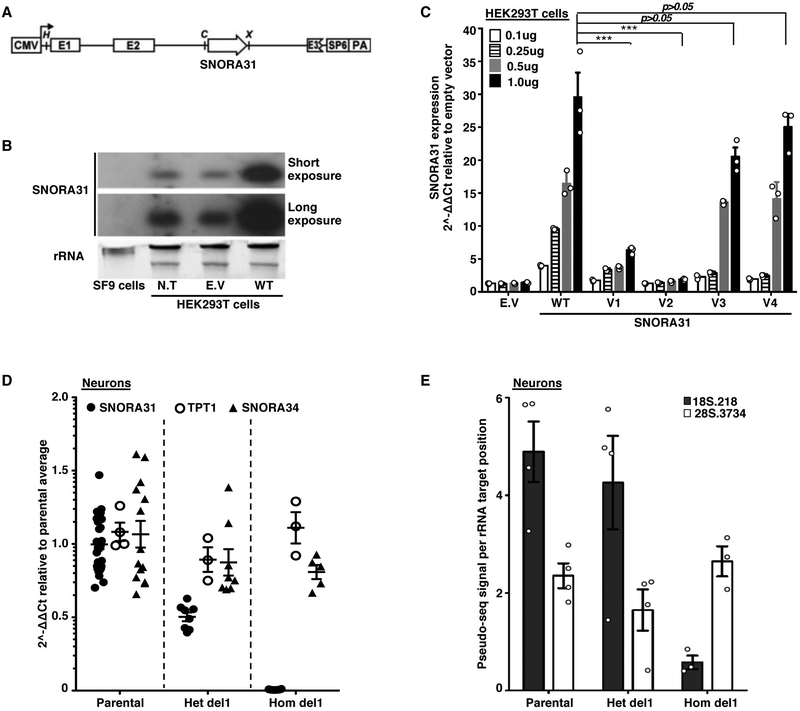
Figure 2. Impaired expression of two SNORA31 variants found in three HSE patients and the loss of a ribosomal modification in SNORA31-null hESC-derived CNS neurons.
A) Schematic diagram of the snoRNA expression vector, into which the wild-type (WT) or mutant sequences of SNORA31 were inserted. Upon expression of the beta-globulin transgene, the immature SNORA31 RNA is spliced at the E2-E3 intron and then processed by the cell machinery into a mature snoRNA molecule. B) Expression of WT snoRNA31 in HEK293T cells, with and without transient transfection with the WT SNORA31 plasmid, as assessed by northern blotting. Insect SF9 cells, which do not express a homolog of SNORA31, were used as a negative control in this experiment. N.T: not transfected. E.V: empty vector. The data are representative of n=2 independent experiments. C) RT-qPCR measurement of snoRNA31 expression in HEK293T cells transfected with the indicated amounts of WT or HSE patient-specific SNORA31 mutant plasmids. The data are expressed relative to SNORD96a expression, with normalization against empty vector transfection. Means and standard deviations from n=3 independent experiments are shown. Mean values of SNORA31 expression levels upon the same amount of WT or mutant plasmid transfection were compared in one-way ANOVA (F=47.51, total df=17) followed by Dunnett’s multiple comparison tests, and the results are shown for the highest amount plasmid transfection group. ***, p<0.001. D) RT-qPCR measurement of relative snoRNA31 expression levels, TPT1 mRNA, and an unrelated snoRNA, SNORA34, in isogenic hESC-derived CNS neurons carrying either a heterozygous deletion (het del1) or a homozygous deletion (hom del1) in SNORA31, or the WT allele (parental). For SNORA31 and SNORA34, data are expressed relative to SNORD96a and normalized relative to the mean value for control cells. For TPT1, the data are expressed relative to GUS mRNA, with normalization relative to the mean value for control cells. The mean and standard deviation from n=3 independent experiments are shown. Each point represents one biological replicate from an independendent experiment, with n=2 (for hom del1), n=3 (het del 1) or n=10 (parental) biological replicates per genotype tested for snoRNA31 expression per experiment. E) Quantification of the pseudo-seq signal at two different rRNA sites in isogenic hESC-derived CNS neurons, either WT for SNORA31 or carrying a heterozygous (het del 1) or homozygous (hom del 1) deletion in SNORA31. Means and standard deviations for n=4 libraries each for the parental and het del1 lines and n=3 libraries for the hom del1 line are shown.