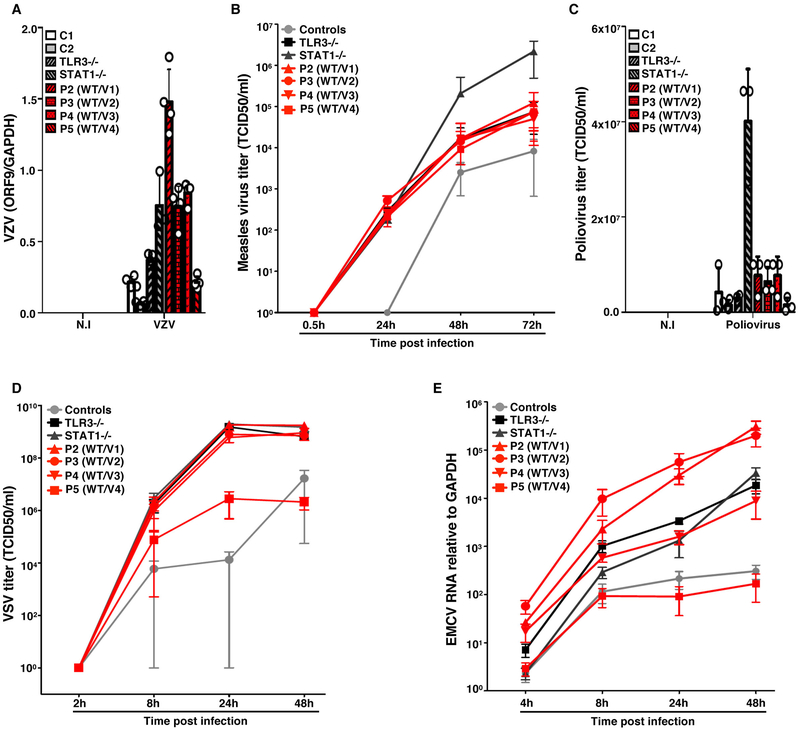
Figure 4. Transcription and propagation of various viruses in the SV40-fibroblasts of patients with SNORA31 mutations.
A) Levels of VZV ORF9 transcript, as determined by RT-qPCR, on SV-40 fibroblasts from patients (P2-P5, red bars with black shadings), a TLR3−/− and a STAT1−/− patient (grey bars with black shadings), and healthy controls (C1 and C2, white and grey bars), 48 h after exposure to VZV-infected MeWo cells (VZV) or MeWo cells that were not infected (N.I). The data are expressed relative to GAPDH expression. Means and standard deviations from n=4 independent experiments are shown. N=1 biological replicate was tested per condition per experiment. B-D) Propagation of measles virus (B), poliovirus (C) and VSV (D), in SV40-fibroblasts from patients and two healthy controls at the indicated times post infection with measles virus at a MOI of 0.5, poliovirus at a MOI of 1, or VSV at a MOI of 1, as assessed by the TCID50 virus titration method. Means and standard deviations from n=3 independent experiments are shown. N=1 biological replicate was tested per condition per experiment. E) Expression levels of the 3D region of the EMCV genome, as measured by RT-qPCR in SV40-fibroblasts from patients (P2-P5), a TLR3−/− and a STAT1−/− patient as susceptible controls, and healthy controls, at the indicated times post infection with EMCV at a MOI of 0.1. The data are expressed relative to GAPDH expression. Means and standard deviations from n=3 independent experiments are shown. N=2 biological replicates were tested per condition per experiment.