Abstract
The present study aimed to analyse the relationship between tumour-infiltrating immune cells (TIICs) and the prognosis of bladder cancer (BC). In the present study, an established computational method (CIBERSORT) was used to analyse the gene expression profile of BC from 409 patients to infer the number of infiltrating immune cells among 22 immune cell subsets. The relationship between each cell type and overall survival (OS) was further analysed. Single-sample GSEA and ESTIMATE algorithms were performed to evaluate the composition of immune microenvironment in each immune cluster. A significant difference in immune cell infiltration between BC and bladder tissue was observed. Increased natural killer and CD8+ T cell infiltration was associated with longer OS, whereas a higher percentage of M0 macrophages among the total immune cells was associated with shorter OS. The number of M0 macrophages increased with increasing BC stage, whereas the percentage of activated memory CD4+ and CD8+ T cells decreased. Patients with BC were divided into three subgroups by hierarchical cluster analysis of immune cells, and each cluster was associated with distinct survival and immune characteristics. The data indicated differences in the cellular composition of TIICs in patients with BC. Moreover, these TIICs were shown to be potential drug targets and reliable prognostic indicators.
Keywords: bladder cancer, tumour-infiltrating immune cell, CIBERSORT, immunotherapy, prognosis prediction
Introduction
Bladder cancer (BC) is the second most common malignancy of the urinary tract and the ninth most frequent cancer; each year 380,000 new cases are diagnosed worldwide and there are ~150,000 deaths reported (1,2). The two types of BC are non-muscle invasive bladder cancer (NMIBC) and muscle invasive bladder cancer (MIBC), of which MIBC is the leading cause of cancer-related mortality in patients with BC (3). Patients with NMIBC exhibit better survival; however, frequent recurrence and eventual progression to MIBC present challenges for successful treatment (4).
Malignant solid tumours consist of tumour cells and interstitial tissue, including stromal cells, vascular endothelial cells, tumour-associated fibroblasts and infiltrating immune cells (5). An increasing number of studies has examined tumour-infiltrating immune cells (TIICs) and suggested that the tumour immune environment influences the prognosis and response to chemotherapy (6,7). In previous studies, immune checkpoint inhibitors have revolutionised the treatment of various human malignant tumours, such as melanoma, classical Hodgkin's lymphoma, non-small cell lung cancer, squamous cell carcinomas and renal cell carcinoma (8–10). The number and distribution of TIICs can be used to divide patients into immunotherapeutic responders and non-responders (11,12).
BC development is highly associated with inflammation and immune cell infiltration. A previous study has also reported the presence of various types of tumour-infiltrating immune cells in BC and signalling pathway interactions between the tumour and TIICs (13). Immunotherapy has been used to treat BC, including intravesical administration of the Bacillus Calmette-Guerin vaccine for treating high-risk NMIBC (14). Numerous immune-based treatments using immune checkpoint inhibitors are under development, including melanoma, non-small cell lung cancer and renal cell carcinoma (15). Incorporating immune cells with current pathology-based stratification may predict the prognosis and therapeutic response in patients with BC.
Immunohistochemistry (IHC) is the most common method for analysing the composition of TIICs. However, IHC can only evaluate a few immune cell types simultaneously (16). The immune response involves highly coordinated interactions between numerous specific cell types. Therefore, to gain insight into the diversity of immune responses, it is necessary to quantify multiple types of TIICs in a large number of patients. CIBERSORT is a systems biology tool that uses extensive deconvolution of gene-expression data and a sophisticated algorithm for in silico quantification of numerous immune cell types in different tumour samples and substrates. The key advantage of CIBERSORT is its high resolving power, which quantifies 22 types of immune cells simultaneously and calculates the relative ratio of each cell type by characterising ~500 marker genes (17). The excellent properties of CIBERSORT were successfully validated by fluorescence-activated cell sorting in several malignant tumours, such as lung cancer, colon cancer and breast cancer, and this tool has been applied to study cellular heterogeneity by many researchers (18–20).
In the present study, CIBERSORT was used to quantify the composition of TIICs in BC and to further explore their relationship with patient survival. This study provides insight into the relationship between TIICs and BC progression.
Materials and methods
Data acquisition
Datasets, including gene expression profiles and corresponding prognosis information from patients with BC, were downloaded from The Cancer Genome Atlas (TCGA; https://cancergenome.nih.gov; uploaded on March 31, 2019). The dataset included 409 patients and 430 samples, of which 19 samples were derived from normal tissues. Samples with P>0.05 were further excluded in the calculation process of CIBERSORT. Finally, 169 samples were included in the analysis. Baseline clinicopathological characteristics were obtained from the datasets including age, sex, tumour grade, T stage, lymph node metastasis, distant metastasis, tumour stage, survival status and survival time.
Evaluation of intratumoural immune cells
The CIBERSORT algorithm was used to calculate the relative percentages of 22 types of TIICs with normalised gene expression data (https://cibersort.stanford.edu) (17). Gene expression datasets were prepared using standard annotation files and then uploaded to the CIBERSORT web portal with the algorithm run using the leukocyte signature matrix (LM22) signature matrix at 1,000 permutations. CIBERSORT calculates a P-value for the deconvolution of each sample using Monte Carlo sampling and provides a confidence value for the results. These TIICs included macrophages (M0, M1 and M2), T cells (CD8+, CD4+ naïve, CD4+ resting memory, CD4+ activated memory, follicular helper, regulatory and γδ), B cells (memory and naïve), resting and activated natural killer (NK) cells, resting mast cells, activated mast cells, resting and activated dendritic cells, monocytes, plasma cells, neutrophils and eosinophils. In the calculation process of CIBERSORT, the type and percentage of each lymphocyte in tumour samples and normal samples can be obtained.
Statistical analysis
Overall survival (OS) was measured from the date of diagnosis to the date of death. Patients were divided into high and low groups according to the median value of the lymphocyte proportion 0.5, then Kaplan-Meier curves were generated to analyse the association between immune cell infiltration and OS, which were then evaluated by the log-rank test. Missing data were processed using the listwise deletion method, and the entire sample was excluded from analysis if any single value was missing. Student's t-test was used to analyse differences in gene expression between tumour and normal tissues. K-clustering was used to determine the appropriate number of clusters. Analysis of variance (ANOVA) was used to test the significance of the difference in the mean of the three groups followed by Tukey's post hoc test. Fold-changes in TIIC percentage were estimated to represent the degrees of expression differences in a volcano plot. R software version 3.5.2(https://www.r-project.org/) was used to conduct all analyses, and all statistical tests were two-sided with P-values <0.05 considered to indicate a statistically significant difference.
Hierarchical cluster analyses
To investigate the relationship between intratumoural immune cells and clinical outcomes, all samples were stratified and clustered to determine whether different patterns of immune cell infiltration could be identified based on the ratios of 22 TIICs. Values for each cell type were rescaled to be between 0 and 1 (the minimum and maximum values observed) to ensure comparability between rare and abundant cell types with low and high proportions. All sample data were stratified and clustered by Ward's method. The possible number of different clusters in the data was explored using a combination of the Elbow method and the Gap statistic. Clusters were divided according to different patterns of immune cell infiltration, and the associations between clusters and clinical outcomes were assessed by Kaplan-Meier curves and compared using log-rank tests. The gene sets of 29 immune markers were defined according to the function of the immune genome (21) and the enrichment level of the gene set in each sample was quantified and sequenced by single-sample geneset enrichment analysis (ssGSEA) (22). The Estimation of STromal and Immune cells in MAlignant Tumors using Expression data (ESTIMATE) was used to evaluate the immune score, stromal cell content and tumour purity of each sample (23).
Results
Patient clinicopathological characteristics
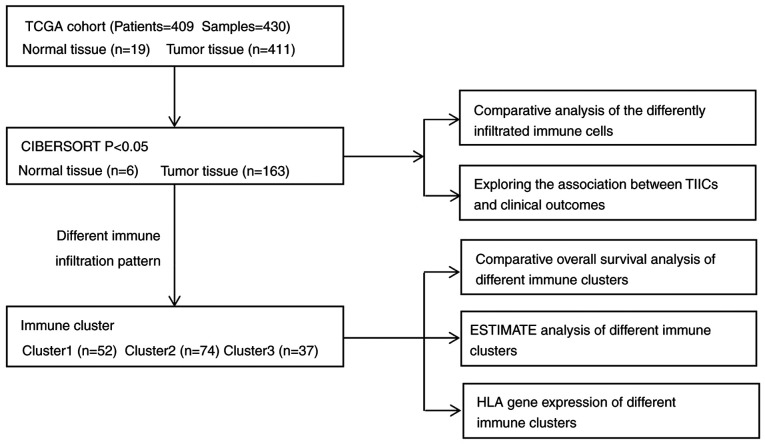
The TCGA dataset included 409 patients with bladder transitional cell papillomas and carcinomas; their general clinicopathological characteristics are listed in Table I. The dataset included 303 men (74.1%) and 106 women (25.9%) with an average age of 68.1 years (range, 34–90 years). Their age, sex, tumour grade, T stage, lymph node metastasis, distant metastasis, clinical stage, survival status and survival duration were recorded at the time of patient admission. Details of the study design and which samples were included at each stage of analysis are illustrated in Fig. 1 as a flowchart. Each tumour sample corresponded to one patient.
Table I.
Baseline characteristics of patients with bladder cancer.
| Clinicopathological characteristic | N (%) |
|---|---|
| Age at operation, years | |
| <60 | 87 (21.3) |
| ≥60 | 322 (78.7) |
| Sex | |
| Male | 303 (74.1) |
| Female | 106 (25.9) |
| Tumour grade | |
| High | 385 (94.1) |
| Low | 21 (5.1) |
| Unknown | 3 (0.7) |
| AJCC stage | |
| I | 2 (0.5) |
| II | 130 (31.8) |
| III | 139 (34.0) |
| IV | 136 (33.3) |
| Unknown | 2 (0.5) |
| Tumour | |
| T1 | 4 (0.9) |
| T2 | 120 (29.3) |
| T3 | 194 (47.4) |
| T4 | 59 (14.4) |
| Unknown | 32 (7.8) |
| Lymph node metastasis | |
| N0 | 237 (57.9) |
| N1-N3 | 131 (32.0) |
| Nx | 41 (10.1) |
| Distant metastasis | |
| M0 | 194 (47.4) |
| M1 | 11 (2.7) |
| Mx | 204 (49.9) |
AJCC, American Joint Committee on Cancer.
Figure 1.
Flowchart of the detailed study design and samples at each stage of the analysis. ESTIMATE, Estimation of STromal and Immune cells in MAlignant Tumors using Expression data; HLA, human leukocyte antigen; TCGA, The Cancer Genome Atlas; TIIC, tumour-infiltrating immune cells.
Distribution of TIICs in bladder cancer
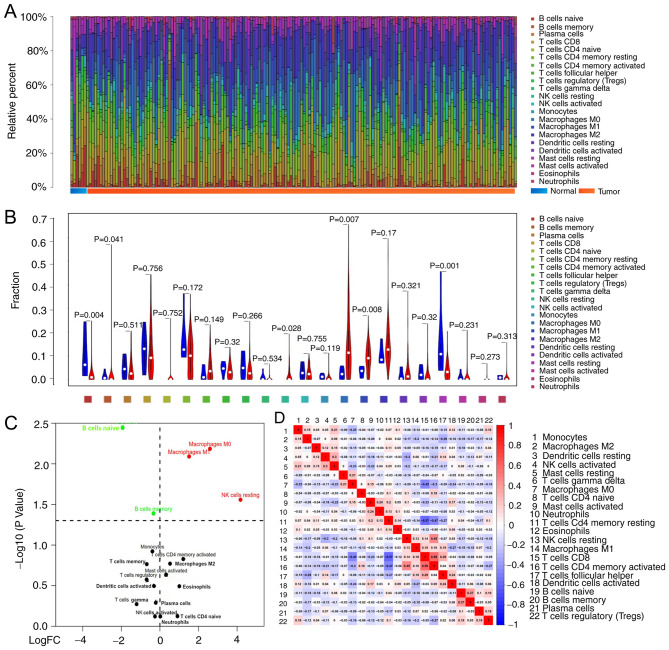
CIBERSORT analysis demonstrated that tumours contained CD8+ T cells (12.3%), resting memory CD4+ T cells (10.2%), M0 macrophages (14.7%) and M2 macrophages (14.9%), whereas tumour infiltrated naïve CD4+ T cells (0.02%), eosinophils (0.02%) and γδ T cells (0.04%) were rare. The composition of TIICs in normal tissue compared to tumour tissue appeared to be different, the sum of relative percentages of TIICs in each tumour sample equal to 1 (Fig. 2A). Normal tissue contained a higher proportion of naïve B cells (P=0.004), memory B cells (P=0.041) and resting mast cells (P=0.001), whereas BC tumours contained relatively higher proportions of resting NK cells (P=0.028), M0 macrophages (P=0.007) and M1 macrophages (P=0.008) (Fig. 2B and C). The number of CD8 T cells was positively correlated with that of CD4 memory activated T cells (r=0.59), but inversely correlated with that of CD4 memory resting T cells (r=0.57); the proportions of other TIIC subpopulations were weakly to moderately correlated (Fig. 2D).
Figure 2.
Differences in immune infiltration between normal and BC tissue. (A) Distribution of immune cell-type fractions in normal tissue and BC. (B) The relative abundance of TIICs in normal (blue) and cancerous tissue (red); comparisons were made using Student's t-test. (C) The differentially infiltrated immune cells in tumoural and normal tissue. The red or green dots in the figure represent TIIC subgroups of high or low expression, respectively. (D) Correlation matrix of 22 immune cell proportions in The Cancer Genome Atlas cohort. BC, bladder cancer; FC, fold change; NK, natural killer; TIIC, tumour-infiltrating immune cells.
Identification of prognostic subsets of TIICs
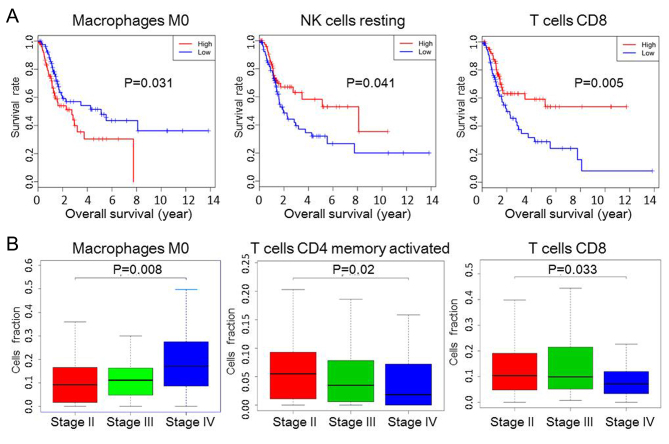
The prognostic value of TIICs was then assessed in BC. The results from Kaplan-Meier curve analysis revealed that a greater number of M0 macrophages (P=0.031) was associated with a shorter OS, whereas increased proportions of resting NK cells (P=0.041) and CD8+ T cells (P=0.005) were associated with prolonged OS (Fig. 3A). Although the composition of other TIICs, such as B cells memory, B cells naive and M1 macrophages, was also different between normal and tumour tissue, they were not statistically significant in determining the prognosis of patients (Fig. S1); similarly, CD8/regulatory T cells (Tregs) ratio was not associated with OS (Fig. S1). The proportion of M0 macrophages increased with increasing tumour stage, whereas the proportion of activated CD4+ memory and CD8+ T cells decreased (Fig. 3B).
Figure 3.
Subgroups of TIIC associated with prognosis in bladder cancer. (A) Kaplan-Meier survival curves of TIIC subpopulations. (B) The relationship between immune cells and tumour stage. NK, natural killer; TIIC, tumour-infiltrating immune cells.
Immune clusters associated with prognosis
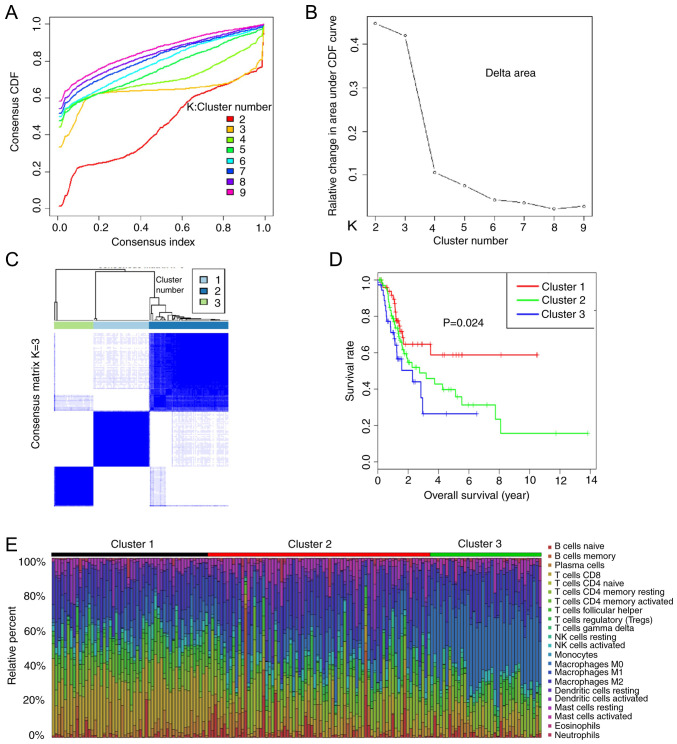
As the TIIC subtypes vary greatly at the individual level, partly reflecting the prognosis, K-means consensus clustering was performed to identify various immune infiltration patterns in patients with bladder cancer (BC). The optimal number of groups is determined by the slope of the curve (Fig. 4A and B). The slope of curves where K ≥3 are no longer significantly different, therefore K=3 is a critical point. The consensus matrix heatmap revealed three clusters (Fig. 4C). Various immune cell percentages in these novel clusters are shown in Fig. 4E, the sum of relative percentages of TIICs in each tumour sample equal to 1. Moreover, the data revealed that different survival patterns were associated with specific clusters (Fig. 4D). Cluster 1, defined by high levels of CD8 T cells and NK cells, showed the best prognosis, whereas cluster 2, with relatively low levels of CD8 T cells and high levels of mast cells showed a worse prognosis than cluster 1. Cluster 3, defined by high levels of M0 macrophages and low levels of CD8 T cells, was associated with poor outcomes. Overall, these findings demonstrated that immune cell infiltration considerably varies in BC, and the presence of different immune cell populations may influence clinical outcomes.
Figure 4.
Association of immune cell clusters with the prognosis of patients with bladder cancer. Combined with (A) elbow method and (B) gap statistics method, the optimal clustering number was calculated and classified into K=3 in a more balanced partition manner. (C) Results obtained from K-means consensus clustering identified three different clusters. (D) Different survival rates of patients in Cluster 1, Cluster 2 and Cluster 3. (E) Distribution of immune cells in three clusters. CDF, cumulative distribution function; NK, natural killer.
Immune characteristics of immune clusters
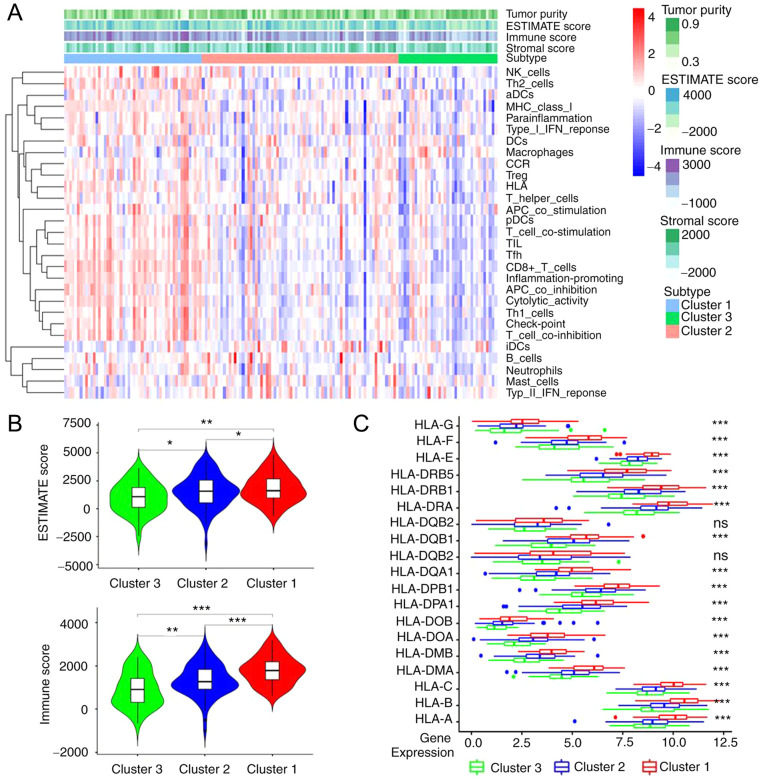
According to the ESTIMATE analysis, the activity or enrichment levels of 29 immune-associated gene sets which represented diverse immune cell types, functions, or pathways in each cancer sample were quantified (Fig. 5A). It was found that found that the ESTIMATE score and the immune score were significantly higher in cluster1 and significantly lower in cluster3, there was a statistical difference among three groups (cluster1 vs. cluster2, cluster2 vs. cluster3 and cluster1 vs. cluster3, all P<0.001; Fig. 5B). Following gene analysis, it was demonstrated that the expression level of most human leukocyte antigen (HLA) genes increased gradually from cluster 3 to cluster 1, there was a statistical difference among three groups (cluster1 vs. cluster2, cluster2 vs. cluster3 and cluster1 vs. cluster3, all P<0.001; Fig. 5C).
Figure 5.
Different immune characteristics of immune clusters. (A) Tumour purity, stromal score and immune score in different immune clusters were evaluated by ESTIMATE analysis. Different colours indicate different percent of purity. The red and blue colour used on the heatmap indicate the high and low relative activity of immune cells, respectively. (B) Comparison of ESTIMATE score and immune score between immune clusters. (C) Comparison of HLA gene expression levels between immune clusters by ANOVA test. *P<0.05, **P<0.01, ***P<0.001. HLA, human leukocyte antigen; ns, not significant.
Discussion
Many types of human tumours have immune cell infiltrates. As cancer development is influenced by host immunity, evaluating the number, phenotype and spatial distribution of immune cells in tumours can provide helpful prognostic information (24). In the present study, the distribution of TIICs in BC based on gene expression data obtained from a TCGA cohort was determined. The data showed that the infiltration patterns of TIICs were significantly different between normal tissue and BC. In addition, analysis of the complex association between TIICs and clinical results revealed the prognostic value of TIICs in BC.
A total of 22 TIICs subsets were quantified in BC, and it was revealed that there are abundant CD8+ T cells, resting memory CD4+ T cells, M0 macrophages and M2 macrophages in BC tumours. The fractions of resting memory CD4+ T cells and M2 macrophages were significantly higher in tumoral tissue, which suggested that they may play an important role in the tumorigenesis of BC, but no association was observed between these two TIIC subsets and clinical outcomes. This reflected the functional heterogeneity of TIIC subgroups during the development of BC.
Tumour-associated macrophages (TAM) can express cytokines and enzymes which could inhibit the recruitment and activation of T cells, thus enhancing resistance to ICIs. Currently, numerous clinical trials are being conducted to inhibit the proliferation or polarization of TAM to enhance the antitumour immune response (25). Macrophages can be divided into M1 (activated) and M2 (alternatively activated) phenotypes according to their functions. These categories represent a range of functional states within the same cell type (26). M0 macrophages, formed from monocytes, have not yet polarised into the M1 or M2 subtypes within the tumour (27). Data from the present study revealed that increased infiltration of M0 macrophages was associated with poor OS and increased tumour stage. Therefore, M0 macrophages may play an important role in BC development, and their differentiation may be therapeutically targetable.
At present, the concept of an ‘immunoscore’ is based on the assessment of T cell subgroups, particularly CD8+ T cells (28). BC has long been thought to interact with the immune system, and many TIICs have been analysed in this tumour type. Lipponen et al (29), observed that CD8+ tumour-infiltrating lymphocytes (TILs) in the tumour stroma were correlated with poor OS. In addition, Horn et al (30) reported that the ratio of Tregs between CD3+ and CD8+ TILs indicates poor prognosis in patients with invasive urothelial cell carcinoma of the bladder. In the stroma, CD8+ TIL density shows a strong positive association with the programmed death-ligand 1 immune cells and the programmed cell death protein 1 TIL density (31). Baras et al (32) reported that the ratio of CD8 to Treg TIL densities was strongly associated with response to cisplatin-based neoadjuvant chemotherapy, supporting the hypothesis that the immune system serves a role in the response of BC to chemotherapy. In the present study, increased levels of CD8+ T cells were associated with prolonged OS, and the proportion of CD8+ T cells decreased with increasing tumour stage. However, the CD8/Treg ratio was not associated with OS, which suggested that factors determining the efficacy of BC are complex and need to be further explored.
NK cells are innate immune cells that are important effector cells in immune surveillance (33). A previous study reported that immune regulation by NK cells could result from a direct interaction between NK cells and T cells, as well as an indirect interaction with antigen-presenting cells, affecting infected cells and the pathogen load (34). In the present study, increased infiltration of resting NK cells was associated with prolonged OS, indicating their protective role in tumour development.
CD4 memory T cells function as helper cells that direct and assist many other cell types and have the potential to perform catalyst functions, increasing the rate of immune protection via multiple different pathways (35). Results from the present study indicated that the proportion of CD4+ activated memory T cells decreased with increasing tumour stage. Furthermore, correlation analysis revealed that the level of CD8 T cells was positively correlated with that of CD4 memory activated T cells but inversely correlated with that of CD4 memory resting T cells. Patients with more CD8 cells in their tumour tissues had better outcomes, which is highly consistent with previous studies (31,32). Therefore, CD4 memory activated T cells may play an important role in tumour progression.
In the current study, patients were divided into different immune clusters according to their tumour immune infiltration patterns, and the OS of different clusters showed significant statistical differences. The composition of tumour microenvironment and the lymphocytes play an important role in the process of antitumour immunity (36). Immune checkpoint inhibitors are an important treatment for advanced stage tumours, its curative effect depended on the immunogenicity of tumour. Compared with cluster 2 and cluster3, cluster 1 had higher levels of CD8 T cells and NK cells. Data from the present study demonstrated that cluster 1 had the highest immune score and the highest expression level of most HLA genes compared with cluster2 and cluster3, which may aid in prolonging the OS of patients and improving the efficacy of immunotherapy (37).
As an emerging technology, the realistic performance of CIBERSORT in BC remains to be verified, and the findings from the present study require validation in a larger external patient cohort before clinical application. Despite these limitations, the findings of this study are informative additions to the existing literature.
In conclusion, this study revealed the relationship between TIICs and BC progression, in addition to the predictive roles of these cells in the outcomes of patients with BC. The diversity of invasive immune cell composition indicated that the immune system may serve a complex role in BC development. Previous evidence suggests that immunotherapy can effectively cure patients with BC, and the selection of personalised treatment for each patient is crucial (15). Data from the present study may improve our understanding of immune responses and provided a valuable resource for studies of immunotherapy.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The current study was supported by The National Natural Science Foundation of China (grant no. 81802723) and The Guangzhou Health and Family Planning Commission Technology Project (grant no. 20191A011104).
Availability of data and materials
The datasets generated and/or analysed during the present study are available in The Cancer Genome Atlas repository (https://cancergenome.nih.gov), and the additional data are available from the corresponding author on reasonable request.
Authors' contributions
MZ and XD conceived and designed the study. YW and HB analysed the data and drafted the manuscript, ZL downloaded the data, directed the statistical analysis and revised the composition of this article. All authors read and approved the final manuscript.
Ethics approval and consent to participate
No applicable.
Patient consent for publication
No applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Malats N, Real FX. Epidemiology of bladder cancer. Hematol Oncol Clin North Am. 2015;29:177–189. doi: 10.1016/j.hoc.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Kaufman DS, Shipley WU, Feldman AS. Bladder cancer. Lancet. 2009;374:239–249. doi: 10.1016/S0140-6736(09)60491-8. [DOI] [PubMed] [Google Scholar]
- 4.Hurst C, Rosenberg J, Knowles M. SnapShot: Bladder cancer. Cancer Cell. 2018;34:350 e1. doi: 10.1016/j.ccell.2018.07.013. [DOI] [PubMed] [Google Scholar]
- 5.Stetler-Stevenson WG. The tumor microenvironment: The connective tissue/tumor cell/host organ system that modulates tumor progression. Connect Tissue Res. 2015;56:343–344. doi: 10.3109/03008207.2015.1086622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fridman WH, Pages F, Sautes-Fridman C, Galon J. The immune contexture in human tumours: Impact on clinical outcome. Nat Rev Cancer. 2012;12:298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 7.Zitvogel L, Galluzzi L, Smyth MJ, Kroemer G. Mechanism of action of conventional and targeted anticancer therapies: Reinstating immunosurveillance. Immunity. 2013;39:74–88. doi: 10.1016/j.immuni.2013.06.014. [DOI] [PubMed] [Google Scholar]
- 8.Branchoux S, Bellera C, Italiano A, Rustand D, Gaudin AF, Rondeau V. Immune-checkpoint inhibitors and candidate surrogate endpoints for overall survival across tumour types a systematic literature review. Crit Rev Oncol Hematol. 2019;137:35–42. doi: 10.1016/j.critrevonc.2019.02.013. [DOI] [PubMed] [Google Scholar]
- 9.Ottaviano M, De Placido S, Ascierto PA. Recent success and limitations of immune checkpoint inhibitors for cancer: A lesson from melanoma. Virchows Arch. 2019;474:421–432. doi: 10.1007/s00428-019-02538-4. [DOI] [PubMed] [Google Scholar]
- 10.Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, Drake CG, Camacho LH, Kauh J, Odunsi K, et al. Safety and activity of anti-PDL1 antibody in patients with advanced cancer. N Engl J Med. 2012;366:2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gnjatic S, Bronte V, Brunet LR, Butler MO, Disis ML, Galon J, Hakansson LG, Hanks BA, Karanikas V, Khleif SN, et al. Identifying baseline immune-related biomarkers to predict clinical outcome of immunotherapy. J Immunother Cancer. 2017;5:44. doi: 10.1186/s40425-017-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masson-Lecomte A, Rava M, Real FX, Hartmann A, Allory Y, Malats N. Inflammatory biomarkers and bladder cancer prognosis: A systematic review. Eur Urol. 2014;66:1078–1091. doi: 10.1016/j.eururo.2014.07.033. [DOI] [PubMed] [Google Scholar]
- 14.Kawai K, Miyazaki J, Joraku A, Nishiyama H, Akaza H. Bacillus Calmette-Guerin (BCG) immunotherapy for bladder cancer: Current understanding and perspectives on engineered BCG vaccine. Cancer Sci. 2013;104:22–27. doi: 10.1111/cas.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim J. Immune checkpoint blockade therapy for bladder cancer treatment. Investig Clin Urol. 2016;57(Suppl 1):S98–S105. doi: 10.4111/icu.2016.57.S1.S98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siska PJ, Johnpulle RAN, Zhou A, Bordeaux J, Kim JY, Dabbas B, Dakappagari N, Rathmell JC, Rathmell WK, Morgans AK, et al. Deep exploration of the immune infiltrate and outcome prediction in testicular cancer by quantitative multiplexed immunohistochemistry and gene expression profiling. Oncoimmunology. 2017;6:e1305535. doi: 10.1080/2162402X.2017.1305535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu X, Wu S, Yang Y, Zhao M, Zhu G, Hou Z. The prognostic landscape of tumor-infiltrating immune cell and immunomodulators in lung cancer. Biomed Pharmacother. 2017;95:55–61. doi: 10.1016/j.biopha.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 19.Xiong Y, Wang K, Zhou H, Peng L, You W, Fu Z. Profiles of immune infiltration in colorectal cancer and their clinical significant: A gene expression-based study. Cancer Med. 2018;7:4496–4508. doi: 10.1002/cam4.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ali HR, Chlon L, Pharoah PD, Markowetz F, Caldas C. Patterns of immune infiltration in breast cancer and their clinical implications: A gene-expression-based retrospective study. PLoS Med. 2016;13:e1002194. doi: 10.1371/journal.pmed.1002194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He Y, Jiang Z, Chen C, Wang X. Classification of triple-negative breast cancers based on Immunogenomic profiling. J Exp Clin Cancer Res. 2018;37:327. doi: 10.1186/s13046-018-1002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hänzelmann S, Castelo R, Guinney J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshihara K, Shahmoradgoli M, Martínez E Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW, Levine DA, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pathria P, Louis TL, Varner JA. Targeting tumor-associated macrophages in cancer. Trends Immunol. 2019;40:310–327. doi: 10.1016/j.it.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 26.Chanmee T, Ontong P, Konno K, Itano N. Tumor-associated macrophages as major players in the tumor microenvironment. Cancers (Basel) 2014;6:1670–1690. doi: 10.3390/cancers6031670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Binnemars-Postma K, Storm G, Prakash J. Nanomedicine strategies to target tumor-associated macrophages. Int J Mol Sci. 2017;18:979. doi: 10.3390/ijms18050979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ascierto PA, Capone M, Urba WJ, Bifulco CB, Botti G, Lugli A, Marincola FM, Ciliberto G, Galon J, Fox BA. The additional facet of immunoscore: Immunoprofiling as a possible predictive tool for cancer treatment. J Transl Med. 2013;11:54. doi: 10.1186/1479-5876-11-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lipponen PK, Eskelinen MJ, Jauhiainen K, Harju E, Terho R. Tumour infiltrating lymphocytes as an independent prognostic factor in transitional cell bladder cancer. Eur J Cancer 29A. 1992:69–75. doi: 10.1016/0959-8049(93)90579-5. [DOI] [PubMed] [Google Scholar]
- 30.Horn T, Laus J, Seitz AK, Maurer T, Schmid SC, Wolf P, Haller B, Winkler M, Retz M, Nawroth R, et al. The prognostic effect of tumour-infiltrating lymphocytic subpopulations in bladder cancer. World J Urol. 2016;34:181–187. doi: 10.1007/s00345-015-1615-3. [DOI] [PubMed] [Google Scholar]
- 31.Wang B, Pan W, Yang M, Yang W, He W, Chen X, Bi J, Jiang N, Huang J, Lin T. Programmed death ligand-1 is associated with tumor infiltrating lymphocytes and poorer survival in urothelial cell carcinoma of the bladder. Cancer Sci. 2019;10:489–498. doi: 10.1111/cas.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baras AS, Drake C, Liu JJ, Gandhi N, Kates M, Hoque MO, Meeker A, Hahn N, Taube JM, Schoenberg MP, et al. The ratio of CD8 to Treg tumor-infiltrating lymphocytes is associated with response to cisplatin-based neoadjuvant chemotherapy in patients with muscle invasive urothelial carcinoma of the bladder. Oncoimmunology. 2016;5:e1134412. doi: 10.1080/2162402X.2015.1134412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pahl J, Cerwenka A. Tricking the balance: NK cells in anti-cancer immunity. Immunobiology. 2017;222:11–20. doi: 10.1016/j.imbio.2015.07.012. [DOI] [PubMed] [Google Scholar]
- 34.Crouse J, Xu HC, Lang PA, Oxenius A. NK cells regulating T cell responses: Mechanisms and outcome. Trends Immunol. 2015;1:49–58. doi: 10.1016/j.it.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 35.Jaigirdar SA, MacLeod MK. Development and function of protective and pathologic memory CD4 T cells. Front Immunol. 2015;6:456. doi: 10.3389/fimmu.2015.00456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tran Janco JM, Lamichhane P, Karyampudi L, Knutson KL. Tumor-infiltrating dendritic cells in cancer pathogenesis. J Immunol. 2015;194:2985–2991. doi: 10.4049/jimmunol.1403134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Darvin P, Toor SM, Sasidharan Nair V, Elkord E. Immune checkpoint inhibitors: Recent progress and potential biomarkers. Exp Mol Med. 2018;50:1–11. doi: 10.1038/s12276-018-0191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analysed during the present study are available in The Cancer Genome Atlas repository (https://cancergenome.nih.gov), and the additional data are available from the corresponding author on reasonable request.