Abstract
The adeno-associated viral vector (AAV) platform has developed into a primary modality for efficient in vivo, and in more limited settings, in vitro or ex vivo gene transfer. Its applications range from a tool for experimental purposes to preclinical and clinical gene therapy. The ability to accurately and reproducibly quantify vector concentration is critical for any of these applications. While several quantification assays are available, here we outline a detailed protocol for the quantification of DNase-I protected vector genomes reliant on the polymerase chain reaction (PCR) as a measure of the active component of the vector, namely its transgene cargo. With the emergence of droplet digital PCR (ddPCR), we provide side-by-side protocols for traditional TaqMan™ real-time, quantitative PCR (qPCR) and ddPCR, as well as comparative data generated with both methods. Lastly, we discuss the importance of the use of surfactant (here, Plutonic® F-68) in the execution of the assay to limit DNA and AAV adherence to various carriers during the titration, particularly at low concentrations. We believe these protocols can lead to reduced variability and increased comparability between AAV studies.
Keywords: AAV, Adeno-associated virus, Vector, Titration, Genome, Quantitative PCR, qPCR, Real-time PCR, Droplet digital PCR, ddPCR
1. Introduction
Recombinant AAV is a complex biologic derived from a nonpathogenic virus that is rendered replication-defective through elimination of all viral open reading frames [1], It is composed of a proteinaceous viral capsid and a single-stranded DNA (ssDNA) vector genome. The vector genome is fully user-defined to deliver transgenic content to a target cell, although the transgene cargo must be flanked by inverted terminal repeats (ITRs), the only element retained from the native virus in the vector genome. In most applications, these ITRs are derived from AAV serotype 2 [2]. These AAV2-based ITR genomes can be packaged into a plethora of natural or engineered AAV capsids [3–5]. Several methods to produce AAV are established that result in vector particles of high concentration and purity, as described in Chapters 3, 7, 19, 21, 22, and 23 [6].
Following production, AAV preparations traditionally undergo various assessments of quantification and qualification, such as vector titration. Titration is a process aimed at quantifying the number of vector particles in a given volume. For AAV, as for many viruses, various assays are available, many of which rely on different principles or measure distinct subpopulations. Specifically, infectious titers determine the concentration of infectious units, which is dependent on the specific conditions of the infectivity assay. Physical titers are analytical measures of the number of physical particles within a set volume. Physical titers for AAV can differ based on the principle that is relied on or the component of the particle that is probed. These include (1) measures of the number of genome containing particles determined by probing for components of the vector genome, (2) measures of the number of assembled capsids as determined by probing for the viral capsid protein, (3) calibrated methods identifying unique physical properties of AAV particles [7–9], and (4) automated imaging methodologies relying on a particle-by-particle assessment and counts. These methods each have their own benefits and limitations, and consequently each may be appropriate depending on the question asked. For example, an assessment of the total amount of assembled capsids is relevant for evaluating safety, and thus in this case measurements of viral capsid protein are preferred.
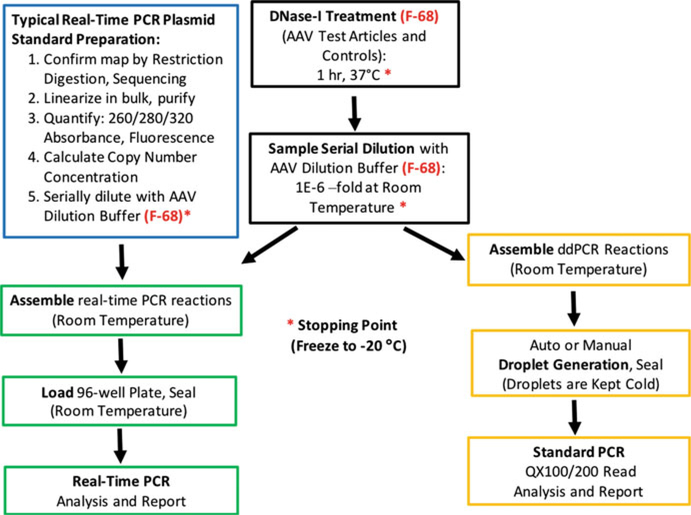
For most applications, however, the active agent in AAV is considered to be the vector genome, irrespective of its use in experimental laboratory or preclinical, translational, or clinical settings. For this reason, most dosing is based on titration methods probing the viral genome, leading to a determination of viral genomes (vg) or genome containing particles (GC) within a given volume. Genome titration is therefore the focus of the protocols below, which leverage the polymerase chain reaction (PCR). Here, we outline and detail a workflow schematically depicted in Fig. 1, leading to the determination of titer using TaqMan™ quantitative PCR (qPCR) or droplet digital PCR (ddPCR) [10]. These methods have been described in various levels of detail previously [11, 12]; however, here we further optimize and compare these different methods. Previous studies have highlighted the potential for large discrepancies among the results of different laboratories using analogous protocols [13]. This protocol aims to provide minute detail to ensure a greater level of standardization across the AAV field.
Fig. 1.
Protocol schematic and workflow
2. Materials
Nuclease-free water (see Note 1).
GeneAmp® 10× PCR Buffer I (Thermo Fisher).
Sheared Salmon Sperm (SSS) DNA (Thermo Fisher) (see Note 2).
Certified DNase- and RNase-free 0.65 mL microcentrifuge tubes, clear.
Certified DNase- and RNase-free 0.65 mL microcentrifuge tubes, colored.
Certified DNase- and RNase-free 1.7 mL microcentrifuge tubes, clear (see Notes 3 and 4).
L20, L200, and L1000 manual micropipettes.
E3–20 and E3–200 LTS electronic pipettes (Rainin Instruments) or electronic equivalent.
Electronic pipette rapid charge stand.
Sterile low-retention filter micropipette tips.
Pipet-Lite™ Multi Pipette L8- or L12–50XLS+ (Rainin Instruments) or electronic equivalent (see Note 5).
Compact PCR Racks for 0.2 mL tubes and strips.
0.2 mL PCR 8-tube strips with attached clear flat caps.
Microcentrifuge tube racks.
Dedicated Microcentrifuge.
Dedicated 8-well PCR tube strip mini centrifuge.
DNase-I: RNase-free with 10× incubation buffer (Roche) (see Note 6).
100× Plutonic® F-68 surfactant (10% solution) (Thermo Fisher) (see Notes 7–9).
10× Plutonic F-68 (1% solution) diluted in nuclease-free water, prepared fresh each time.
100× SSS DNA (200 ng/μL solution) in 1× PCRBuffer I and 0.1% Plutonic® F-68.
10× SSS DNA (20 ng/μL solution) in lx PCR Buffer I and 0.1% Plutonic® F-68.
AAV sample dilution buffer: lx PCR Buffer I, 2 ng/μL SSS DNA, 0.1% Plutonic® F-68 (see Note 10).
Blank buffer (Mock negative control): PBS, 35 mM NaCl, 0.001% Plutonic F-68, or nuclease-free water.
AAV validation sample for quantitative positive control (see Note 26).
DNase-I linear plasmid control (1E+8 copies/5 μL), serially diluted from stock using AAV sample dilution buffer.
AAV experimental samples.
Forward sequence detection primer working stock solution: 9 μM for ddPCR, 3 μM for qPCR (see Notes 11 and 12).
Reverse sequence detection primer working stock solution: 9 μM for ddPCR, 3 μM for qPCR (see Notes 11 and 12).
TaqMan™ fluorescent probe working stock solution: 2.5 pM for ddPCR, 2 μM for qPCR (see Note 13).
Applied Biosystems SDS7500 Sequence Detector (Thermo Fisher), or equivalent.
Restriction enzymes with reaction buffer.
Agarose gel with Ethidium Bromide or equivalent.
QIAquick PCR Purification Kit (QIAGEN) with EB Buffer or Plasmid Purification Kit.
Linearized plasmid standard set, 1E+8 to 1E+1 copies/5 pL, serially diluted from stock in AAV dilution buffer.
Calibrated spectrophotometer (see Note 14).
Qubit™ Fluorometer (Invitrogen), or equivalent.
Qubit™ dsDNA HS Assay Kit (Invitrogen), or equivalent.
2× ddPCR™ Supermix for Probes, no dUTP (Bio-Rad).
2× TaqMan™ Universal Master Mix for qPCR (Thermo Fisher).
Auto DG™ Automated Droplet Generator (Bio-Rad) [14].
Automated Droplet Generation Oil for Probes (Bio-Rad).
DG32™ AutoDG™ Cartridges (Bio-Rad).
AutoDG™ droplet cooling box.
QX100™or QX200™ ddPCR System (Droplet Reader) and QuantaSoft™ Software (Bio-Rad) [15, 16].
C1000 Touch™ Thermocycler (Bio-Rad), or equivalent.
PX1 PCR Plate Sealer and corresponding Plate Support Block (Bio-Rad).
Pierceable Foil Heat Seal (Bio-Rad).
ddPCR 96-well plates (Bio-Rad) or equivalent.
Optical 96-Well Reaction Plate for qPCR.
Optical Adhesive Film Kit for qPCR (Thermo Fisher).
Splash-Free 96-Well Base (Thermo Fisher).
TempPlate® sealing film, non-sterile (USA Scientific).
Corning™ Costar™ 50 mL Sterile Disposable Reagent Reservoirs (Fisher Scientific), or equivalent.
Powder-free Nitrile Gloves.
Clean Lab Coat.
BSL-2 Biological Safety Cabinet with UV light.
15 mL conical tubes.
Kimwipes™ Delicate Task Wipers, or equivalent.
Paper towels.
Wet ice and ice bucket.
Timer.
Vortex Mixer.
Lysol.
70% Ethanol.
Lab notebook and pen.
Adhesive tape.
Color markers, fine and extra-fine point.
3. Methods
3.1. Preparation of the Work Area (See Note 15)
Work in a biosafety cabinet or HEPA-filtered PCR station with UV light capability, preferably a dedicated unit for quantification of high copy number targets.
Wear gloves at all times and change them often, particularly after leaving (and returning to) the work area.
Apply a liberal amount of Lysol or equivalent to the work surface area inside the cabinet and wipe it clean with a paper towel. Do the same again with 70% Ethanol.
Apply a small amount of Lysol or equivalent to a paper towel and wipe clean the microcentrifuge tube racks that will be used. Repeat with 70% Ethanol. Place the clean racks well inside the clean biosafety cabinet.
Apply a liberal amount of Lysol or equivalent to pipette shafts (manual or electronic) over a biohazard trash bag. Repeat with 70% ethanol and wipe the pipette shafts dry with a lint-free Kimwipes™. Place the pipettes in the clean biohazard cabinet.
Protect the LED screen of electronic pipettes with aluminum foil while the units are exposed to UV light. Exposure to UV light will crack LED screens over time (Fig. 2).
Turn on the UV light for 15 min (see Note 16).
Fig. 2.
Protect the LED screens of electronic pipettes from UV light damage
3.2. Preparation of the Real-Time PCR (qPCR) Standard
Prepare the real-time PCR standard in a biological safety cabinet or PCR station, rather than on the bench, where reagents or standards are more likely to become contaminated. Use dedicated reagents.
The standard is usually a cis-plasmid (i.e., ITR-flanked transgene construct) used in the manufacturing of the virus. There are two conditions that need to be met in advance of standard preparation: the plasmid must be a clean, clonally pure species (OD 260/280 between 1.80 and 1.90) and an accurate electronic map must be available (see Note 17).
Confirm the clonal purity of the plasmid by performing an analytical restriction digestion assay of the DNA. Ideally, the assay will target both the recombinant AAV genome embedded in the plasmid and the plasmid backbone. Set up several reactions and choose enzymes that will target several components of the recombinant genome.
Linearize the plasmid with 10–20 units of enzyme in the appropriate reaction buffer, preferably overnight. Digest 10 pg of plasmid per 50 μL reaction. For long-term use, 40 pg of linearized plasmid will last for months or years if properly stored (see Note 18).
Use 1% of each reaction (0.5 μL) to verify the efficiency of linearization in separate wells of an agarose gel. There must be a single band per lane in the gel. Run appropriate controls, as needed, side by side.
Clean up the linearization reactions using QIAGEN’s QIAquick® PCR Purification Kit (or a plasmid purification kit). Elute twice with 50 μL of EB Buffer in a DNase- and KNase-flee 1.7 mL tube. Pool the eluates into a single tube and label (see Note 19).
Quantify the pooled linearized plasmid using spectrophotometry or fluorometry, but preferably both (see Note 20).
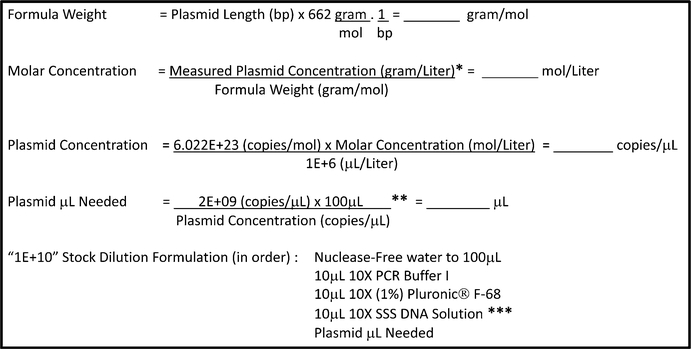
Determine the copy number concentration of the linearized plasmid and the volume needed for the first standard dilution (Fig. 3). Round off all the estimated volumes for pipetting to two decimal points. A final concentration of 0.1% Plutonic® F-68 must be included at the outset in the first dilution of the standard (lE+10 or 1E+09 copies per 5 μL, depending on the yields from steps 6 and 7). The “1E+10” and/or the “1E+09” dilution constitute the working standard stock solutions of the linearized plasmid (see Notes 21–23).
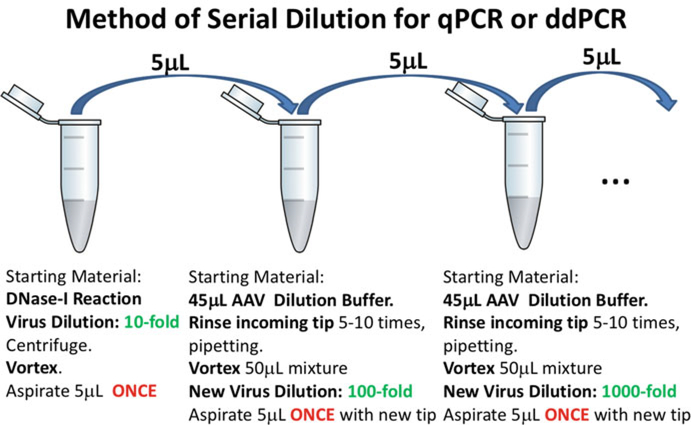
Serially dilute the working stock oflinearized plasmid standard (lE+10 or 1E+09 copies/5 μL) in tenfold dilutions to 1E+01 copies/5 μL in AAV sample dilution buffer. Vigorously vortex each previous dilution before moving on to the next, and always rinse the tip carrying the plasmid by pipetting up and down 5–10 times after addition of plasmid to diluent (Fig. 4) (see Note 24).
Discard 10 μL from the “1E+01 copies/5 μL” dilution to keep the volumes of the standard set equal across all tubes. The “1E+08 copies/5 μL” through the “1E+01 copies/5 μL” dilutions comprise the standard set to be used downstream for quantification purposes in real-time PCR.
From the “1E+09” working stock dilution, prepare an additional “1E+08 copies/5 μL” dilution of the linear standard in AAV dilution buffer to serve as the DNase-I plasmid control in all future quantification runs. Store the linearized plasmid standard dilutions and stocks at −20 °C (see Note 25).
Fig. 3.
Workflow for determining linear plasmid copy number and dilution. Calculate the copy number concentration of the linearized plasmid and the volume needed for a dilution of 2E+9 copies per μL (1E+10 copies per 5 μL), based on the known length of the plasmid, “Plasmid Length (bp),” and the mass concentration of the linearized plasmid as determined in Subheading 3.2, steps 6 and 7, “measured mass concentration (g).” Choose pipettes that maximize the accuracy and precision of reagent delivery. *See Subheading 3.2, steps 6 and 7 and Notes 19 and 20. **The final concentration of the stock linear plasmid dilution may be adjusted according to the linear plasmid yields obtained. For example, this value may be reduced from 2E+09 to 2E+08 copies/μL to create a stock of 1E+09 copies/5 μL, rather than 1E+10 copies/ 5 μL, if the yield of linear plasmid is low. ***See Subheading 2 for exact formulation
Fig. 4.
Serial dilution for MV quantification. Follow the same protocol whether diluting virus or plasmid
3.3. Sample Preparation: DNase-I Treatment
Prepare the surface area of a dedicated biosafety cabinet as specified in Subheading 3.1, above. In addition, clean a compact PCR rack with Lysol and 70% ethanol and add to the work space.
Wear a lab coat and gloves for this procedure. Change gloves often, especially after leaving (and returning to) the work area.
Thaw and equilibrate the AAV sample dilution buffer to room temperature.
Pulse-centrifuge the AAV test samples and controls to collect them at the bottom of the tube (see Note 26).
Transfer as many 0.2 mL 8-well tube strips as needed to the compact PCR rack in the biohazard cabinet for the DNase-I treatment step. Close all the lids. The DNase-I incubation will be carried out in a programmable thermocycler that can accommodate the strips.
Organize the AAV test samples in ascending order followed by the controls and assign a number to them. Label the flat caps of the 0.2 mL tube strips in advance with the assigned number (see Note 27).
The DNase-I treatment step will be the first tenfold dilution of the AAV test samples and controls (see steps 3 and 4 in Subheading 3.4). All dilutions made going forward will be tenfold serial dilutions.
Prepare 10× Pluronic® F-68 (1%) reagent (5 pL per reaction). Make enough for all samples and controls, with sufficient extra volume to offset pipetting errors.
-
Prepare the DNase-I reaction mix for all samples and controls in a 1.7 mL microcentrifuge tube as follows (45 μL per reaction):
Reagent Volume per sample Nuclease-free Water 33 μL 10× DNase Buffer 5 μL 10× (1%) Pluronic F-68 5 μL DNase I, RNase-free 2 μL (20 units) Sample (AAV) 5 μL Total Volume 50 μL Multiply the “volume per sample” from above by the number of samples (AAV test samples and controls) to determine the total volume of reagents that will be needed for this step. Include two additional volumes in the calculation to offset pipetting errors (see Note 28).
Before adding DNase-I to the reaction mix, dispense 43 μL of the DNase-free reaction mix into the DNase-I plasmid control tube that will not include the enzyme. Bring to 45 μL with 2 μL of nuclease-free water (see Note 29).
Add the total volume of DNase-I needed for the remaining reactions to the 1.7 mL tube with the reaction mix, plus two extra volumes (see Note 30).
Invert the (complete) DNase-I reaction mix several times and only gently vortex it once. Dispense 45 μL to the remaining tubes in the strips, one at a time, using the multi-dispense function of an electronic 200 μL pipette. Immediately close the lid of each tube after dispensing the mix before moving on to the next one. Centrifuge the strips.
Add the test samples and controls to the DNase-I reactions (see Note 21). Vigorously vortex one sample or control at a time and add 5 μL to the corresponding reaction in the 8-tube strips in the compact PCRrack. Pipet up and down 5–10 times in the reaction mix to rinse the pipette tip (see Note 31).
Place the tube strips in the C1000 Touch thermocycler block and incubate the DNase-I reactions at 37 °C for 1 h (see Note 32).
Return all original samples and controls to the refrigerator or freezer (see Note 33).
3.4. Sample Preparation: Serial Dilution
Prepare the work area as described in Subheading 3.1.
Allow the DNase-I reactions to equilibrate to room temperature after taking them out of the thermocycler. Centrifuge the strips and vigorously vortex to mix.
AAV test samples, Mock controls, and Validation controls will be serially diluted 100,000-fold more (1,000,000-fold total) with AAV sample dilution buffer. Prepare five 0.65 mL microcentrifuge tubes for each sample or control for the serial dilution step.
The DNase-I plasmid controls will be diluted 10,000-fold more (100,000-fold total) with AAV sample dilution buffer. Prepare four 0.65 mL tubes for each plasmid control for the serial dilution step.
Set up the 0.65 mL microcentrifuge tubes so that they follow the same organization as the samples in the 0.2 mL strip tubes along the bottom length of a microcentrifuge tube rack(s).
For brevity, label only the last 0.65 mL tube in each dilution series (the dilution that will be used to deliver the sample to the PCR reaction).
Dispense 45 μL of the room temperature-equilibrated AAV sample dilution buffer to each 0.65 mL dilution tube using the multi dispense mode of an electronic 200 μL pipette (see Note 34).
Serially dilute each DNase-I reaction (Fig. 4).
Because most of the 0.65 mL tubes in the rack will not be labeled, use the empty slots in the microcentrifuge tube racks to track your progress, avoid confusion during pipetting, and minimize errors due to slapped tubes. Move each tube up a slot in the rack after it has been used to track progress until completion (see Note 27).
Place the most dilute samples (the labeled tubes) on ice until the next step. Repeat until all the samples or controls are diluted. Discard all unlabeled tubes. Return the initial DNase-I reactions to the freezer and do not discard them until results are obtained (see Notes 35 and 36).
For ddPCR, proceed to Subheadings 3.5 and 3.6. For qPCR, proceed to Subheading 3.7.
3.5. ddPCR Reaction Setup and Droplet Generation (See Note 37)
Prepare the work area as described in Subheading 3.1. Only one clean microcentrifuge tube rack will be needed to hold the serially diluted samples and controls, as well as the bulk reaction mixture preparations. Also include a splash-free 96-well base to hold a Bio-Rad 96-well plate (or equivalent) where the reactions will be ultimately assembled.
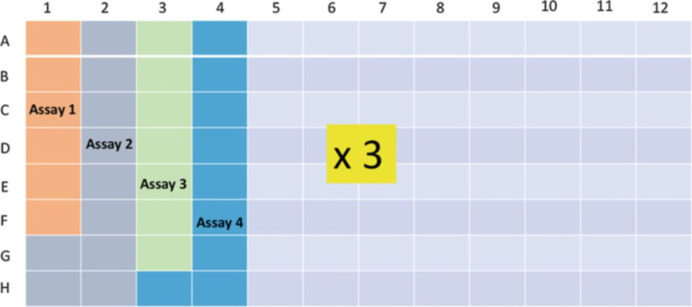
While the interior of the biohazard cabinet is exposed to UV light, create a map of the 96-well plate indicating the position of all samples and controls in each TaqMan™ assay that will be included (Fig. 5).
-
Set aside a 1.7 mL microcentrifuge tube for each TaqMan™ assay. Prepare all ddPCR reactions for each assay in a master mix as follows (18 μL per reaction):
Reagent Vol. per reaction (μL) 2× ddPCR™ Supermix for Probes 10 Forward Primer (9 μM) 2 Reverse Primer (9 μM) 2 TaqMan™ Fluorescent Probe (2.5 μM) 2 Nuclease-free water 2 Sample; (AAV or control) 2 Final Volume 20 Calculate the number of reactions that are needed to quantify all samples and controls in triplicate. Include additional 6–8 reactions per assay to offset pipetting error. The suggested sample order for each TaqMan™ assay is as follows: Test samples (in ascending order), DNase I (−) Plasmid Control, DNase I (+) Plasmid Control, Mock Control, AAV Validation Control, and NTC (No Template Control). A maximum of 32 unique samples may be analyzed in triplicate per plate (see Note 38).
Mix the equilibrated 2× ddPCR Supermix (No dUTP) by inversion and prepare the ddPCR master mix(es) according to the instructions above. Vortex gently to mix.
Carefully transfer 18 μL of master mix to the bottom of each reaction well following the map generated in step 2; use the multi-dispense function of a 200 pL electronic pipette (single or multichannel) (see Note 39).
Transfer 2 μL of each DNA sample or control to the side of each designated well using the multi-dispense function of a p20 electronic pipette and following the map generated in step 2 (see Note 21). Open one tube at a time while dispensing and keep tube lids closed when not in use.
After completion, seal the plate with lightly applied TempPlate® sealing film (see Note 40).
Start the heat-sealing program in a PX1 PCR Plate Sealer (180 °C for 5 s) and wait for it to reach maximum temperature. The program will not be able to start until maximum temperature is reached. The program will hold until the start button is pressed in step 11.
Spin down the plate in a dedicated PCR 96-well plate centtifuge at 226× g for 30 s.
Transfer the plate to the plate support block in the PX1 PCR Plate Sealer.
Carefully remove the TempPlate® sealing film while holding the plate down, and replace it with a sheet of pierceable foil heat seal (with red marker line up toward the top row of the plate). Close the PX1 Plate Sealer and start the heat-sealing program that has been holding since step 8.
Remove the plate and briefly vortex to mix the reactions. Centrifuge the plate again.
Program the AutoDG™ by matching the position and number of columns on the plate that hold reactions to their exact location in the droplet generation map provided by the instrument. The AutoDG™ will calculate the amount of materials and the volume of droplet generation oil that will be needed for the plate you have programmed in the system.
Set the plate inside the AutoDG™ system in the designated location for droplet generation.
Populate the AutoDG™ with the necessary materials requested by the instrument to complete the task: DG32™cartridges (up to three), boxes of tips (up to two), droplet cooling box, and a new 96-well plate to contain the newly formed droplets.
Once everything is in place, lower the AutoDG™ door hatch and start the instrument. For a full plate of reactions, the AutoDG™ may take up to 45 min to complete.
Droplet formation is affected by temperature. Ideally, the room where the AutoDG™ is kept should be between 20 and 25 °C. If the temperature is lower than this, the AutoDG™ may generate an error during the execution of the droplet generation program. To date, in our experience such errors have not impacted results (see Notes 41 and 42).
Fig. 5.
Schematic view of a 96-well plate layout for the Bio-Rad QX200® Quanta Soft™ ddPCR software. Sample and TaqMan™ assay locations are mapped in advance to assist in the calculation of reagent volumes and determination of loading strategy
3.6. Droplet Digital PCR and Plate Reading
Droplets are collected in a new plate over a cooling block. The plate must be heat-sealed promptly without disrupting the droplets, then transferred to a thermocycler for PCR. This should be done within an hour of droplet formation. Keep the plate on the cooling box at all times. The droplets are fragile at this stage, but will be stabilized during PCR.
-
Use the following PCR Reaction conditions:
- 95 °C, 10 min
- 40 cycles of 94 °C, 30 s and 60 °C, 1 min
- 98 °C, 10 min
- 4 °C, indefinitely
The PCRramp rate should be between 2 °C/3 °C per s. Set the final reaction volume to 40 μL (see Note 43).
Open the QuantaSoft™ application and use the map generated in step 2 of Subheading 3.5 to create a droplet readout file. Assign the quantification protocol to be used (ABS), supermix type, sample names, and TaqMan™ assay to all wells.
Insert the plate in the QX200™ reader and initiate the droplet count. It will take an additional 2.5 h to read a full plate.
After completion, click OK and analyze the run. Select the ID amplitude chart from the available options under the “Analyze” tab, and manually assign a threshold to all wells with the same TaqMan™ assay. The “Analyze” tab allows the operator to choose whether to assign thresholds on an individual reaction basis or as a group. Set the threshold value such that it falls just above the negative droplets at the bottom of the chart. The software will automatically calculate a target concentration per μL of reaction.
After the threshold values have been assigned, save the document and export the .csv file to a USB drive for further analysis and final calculations.
- From the “Concentration” column in the .csv file (Copies/ μL), calculate AAV Titer (Genome Copies/mL) using the following formula (see Note 44):
- For the plasmid DNase-I controls, data should be reported as “Copies per DNase-I reaction” rather than as a concentration, in order to determine the recovery of total plasmid input. Calculate using the following formula (see Note 45):
- The background signal from the “No Template Control” (NTC) wells should also be reported as “Copies per reaction.” Calculate using the following formula:
Generate a report from the .csv data and save the file as an Excel sheet or workbook (Table 1). Distribute the report to the AAV production team and quality assurance supervisors for review (see Note 46).
Table 1.
Example ddPCR Report
| Sample ID | Titer #1 | Titer#2 | Titer #3 | Average | Stnd Dev | Notes |
|---|---|---|---|---|---|---|
| Sample A TFF | 2.52E+12 | 2.43E+12 | 2.14E+12 | 2.36E+12 | 1.99E+11 | |
| Sample A Final | 2.45E+13 | 2.17E+13 | 2.03E+13 | 2.22E+13 | 2.16E+12 | |
| 1E8 IE-5 (Plasmid DNase-I Control) | 9.25E+07 | 8.85E+07 | 6.35E+07 | 8.15E+07 | 1.57E+07 | Copies per reaction |
| 1E8D IE 5 (Plasmid DNase-I Control) | 6.00E+05 | 1.55E+06 | 1.50E+06 | 1.22E+06 | 5.35E+05 | Copies per reaction |
| M/B (Mock Control) | 3.70E+09 | 2.30E+09 | 2.40E+09 | 2.80E+09 | 7.81E+08 | |
| XXXX (Validation Control) | 1.20E+13 | 1.08E+13 | 9.71E+12 | 1.08E+13 | 1.16E+12 | |
| NTC | 2.40E+00 | 8.00E+00 | 3.60E+00 | 4.67E+00 | 2.95E+00 | Copies per reaction |
In-process samples (TFF) and final concentrates are routinely quantified in our labs, as well as crude lysates. The latter require less dilution. The validation control samples are reproducible to within 10–30% with very low background each run (<0.01%). Intra-assay reproducibility is within 10–30% among different operators
3.7. Real-Time PCR (qPCR) (See Note 47)
Prepare the work area as described in Subheading 3.1.
Thaw the qPCR standards.
Thaw the working TaqMan™ assay stock for qPCRin advance. The oligos should amplify the same target sequence that was used to quantify test samples and controls by ddPCR.
Remove the 2× TaqMan™ Universal master mix from storage. Equilibrate all reagents to room temperature as before.
Prepare 0.65 mL colored tubes for the real-time PCR reactions: one for NTC, eight for standards, and one for each test sample or control.
All real-time PCR reactions will be carried out in a 50 μL final reaction volume in triplicates for each standard, sample, or control (see Note 48).
- Prepare the real-time PCR master mix as follows:
Reagent Vol. per reaction (μL) TaqMan™ Universal PCR Mix (2×) 26.25 Forward Primer (3 μM) 5.25 Reverse Primer (3 μM) 5.25 Fluorescent Probe (2 μM) 5.25 Nuclease-free water 5.25 Test Sample or control 5.25 Total 52.50 Multiply the volume of each reagent by the total number of NTCs, standards, and samples and controls in triplicate. Include an additional two volumes to offset pipetting errors. Prepare the master mix in a 1.7 mL microcentrifuge tube or 15 mL conical tube. Mix by vortexing.
Take the same precautions for qPCR as for ddPCR when dispensing reagents and samples. Keep all lids closed (tubes, tip boxes) when not in use. Avoid aerosol formation by limiting blowouts and change gloves often, especially after leaving (and returning to) the work area.
Transfer 141.75 μL of PCR master mix to each 0.65 mL tube. Add 15.75 μL of nuclease-free water to the NTC reaction, then add 15.75 μL of each standard, sample, or control DNA to the predesignated tube. Mix each tube by vortexing.
Dispense 3 × 50 μL of the reactions to designated wells in the qPCR plate using the multi-dispense function of an electronic pipette, starting with the standards, followed by samples or controls, and ending with the NTC.
Seal the plate with optical film and spin it down at 226 × g for 30 s.
Set up a dedicated file for the run using the SDS software and load the plate into the qPCR instrument.
- Use the following PCR Reaction conditions:
- (a) 10 min at 95 °C
- (b) 40 cycles of 95 °C, 15 s and 60 °C, 1 min
Allow the instrument to establish the threshold automatically.
-
Calculate the final titers (GC/mL) using the following formulas.
For ssAAV and mock control (see Note 49):For scAAV:For plasmid controls:For NTC, no “Qty Mean” is provided in the SDS7500 system. The Ct values are used as an estimate of background signal. Alternatively, a “Qty Mean” value may be calculated independently from the standard curve data if desired.
3.8. Limitations of the Protocol
This protocol relies on the denaturation temperature and incubation time of the PCR to break open vector capsids and make AAV genomes therein accessible for quantification. AAV serotypes are differentially stable and we may well be underestimating the titers of the most stable capsids [8, 17].
Some AAV genomes or target sequences therein may be refractory to amplification despite our best efforts for efficient quantification, due to entrenched secondary structures, potential methylation of CpG islands at oligonucleotide binding sites, repeat sequences, or our inability to adequately visualize the AAV genome in solution [18]. Unknown, distant interactions may lead to incorrect placement of TaqMan™ assay oligonucleotides. In this case, the combination of qPCR and ddPCR for the same target sequence may yield valuable insights regarding amplification efficiency for the improvement of AAV genome quantification by qPCR. Alternatively, methods that rely on AAV capsid (protein) quantification (semiquantitative SDS-PAGE, ELISA, and AUC) are needed to complement AAV genome quantification by PCR and achieve an analytical understanding of vector lot composition. In the absence of empty capsids and spurious DNA encapsidation products, the successful assembly of recombinant virions is expected to yield a 1:1 capsid-to-genome ratio, and this ratio should serve as a benchmark for the appraisal of quantification methods and packaging efficiency of the recombinant vectors on a lot-tolot basis. The use of surfactant during genome quantification is crucial for this assessment.
Limitations may be imposed by the biology of AAV production itself, where heterogeneity of packaged genomes can make it difficult to confidently assess the concentration of the genome sequence ofinterest [19]. In this case, neither PCRmethod nor AAV capsid quantification can reliably assess the integrity of AAV preps; modifications to the biology of AAV production must be implemented for these highly specialized constructs with subsequent evaluation by next generation sequencing (see Chapter 5).
3.9. Conclusion
Our data indicate that the adaptation of Plutonic® F-68 from an AAV manufacturing setting [20] to vector quantification work markedly increases quantification accuracy, and moves us closer to a true representation of vector concentrations in recombinant AAV preparations. It is now possible to offer critical quality control support to vector manufacturing facilities from the perspective of genome quantification, as it is possible to arithmetically track AAV genome content in a highly specific manner and with minimal loss of sample. This detailed protocol for AAV quantification by qPCR and ddPCR has been presented in the spirit of current guidelines for publication [21, 22] and to address concerns by the gene therapy community [23]. Our data also suggest that ddPCR and qPCR are equivalent technologies for accurate AAV quantification if the physical properties of the viral capsid are observed. Such considerations may need to be extended to other quantitative assays that are currently hindered by the adherence of the AAV capsid to plastic surfaces, most notably the semiquantitative SDS-PAGE assay [24], which together with PCR provide an overall view of AAV genome packaging efficiency and product quality. Further optimization work is in progress.
Fig. 6.
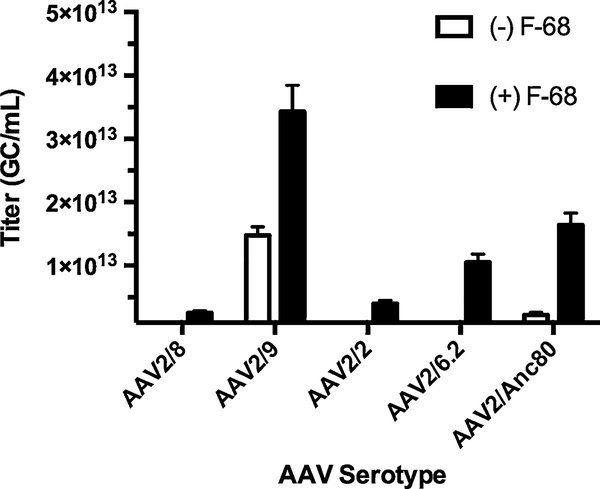
ddPCR titration is improved by inclusion of Pluronic® F-68 according to AAV serotype. The addition of F-68 during AAV guantification confers to a significant increase in signal that improves vector guantification. The degree of gain ranges from modest (two- to threefold) for AAV2/9 and AAV2/8, to large (five- to tenfold) for AAV2/Anc80 and AAV2/2. The same TagMan™ assay was used for these measurements, and all five serotypes carried the same single-stranded genome. Based on these observations, it appears that AAV capsids differentially attach to plastic
Fig. 7.
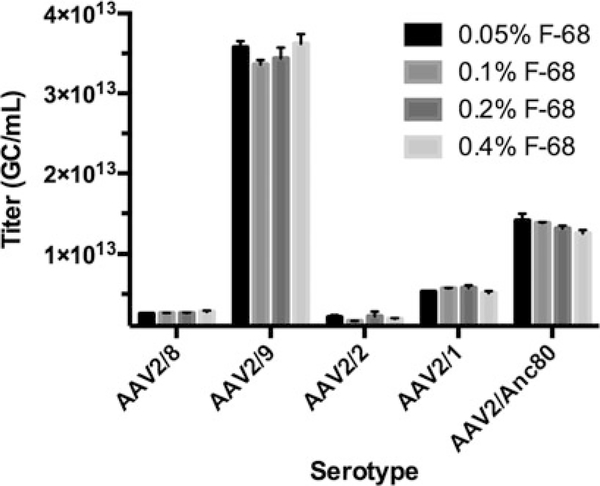
Pluronic® F-68 optimization for ddPCR. Increasing the amount of F-68 does not significantly improve AAV titration of several serotypes. The same TagMan™ assay was used for all measurements, and all vectors carried the same single-stranded genome. In light of these results, it was decided to abide by the manufacturer’s recommendations for future measurements, and to use a final F-68 concentration of 0.1% (1×) for untested AAV capsid variants
Fig. 8.
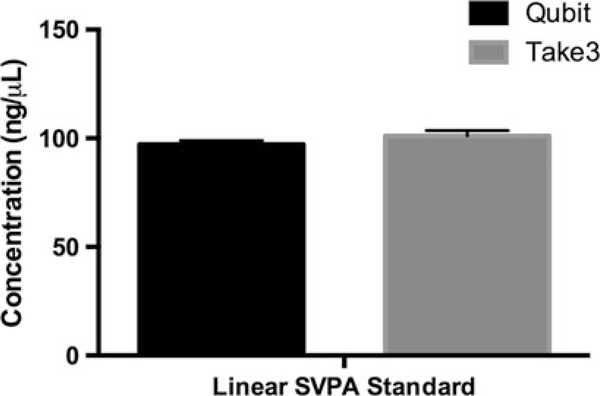
Quantification of a linearized qPCR standard plasmid. Fluorometry (Qubit™) was compared against Spectrophotometry (BioTek® Take3™). When the DNA being measured is clonally pure and clean, these two types of measurements closely agree. Fluorometry measurements may be lower than spectrophotometer O.D. measurements, because the former is highly specific for double-stranded DNA
Fig. 9.
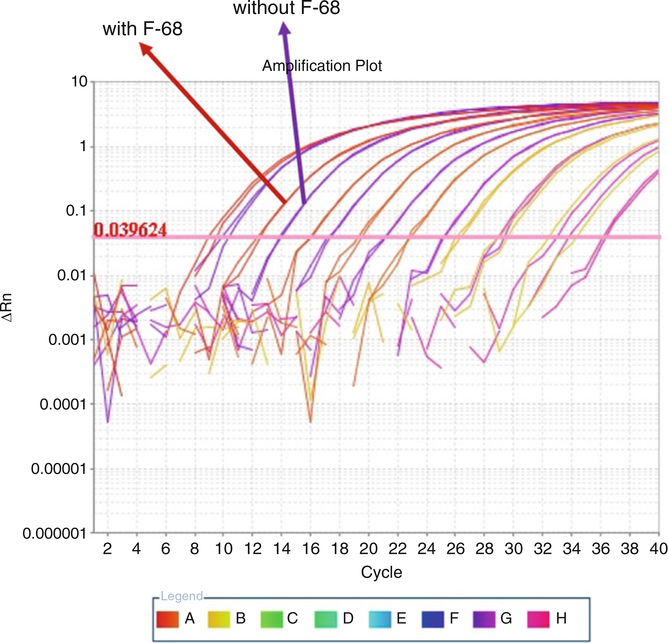
qPCR amplification plot showing the effect of 0.1% Pluronic® F-68 on qPCR standard performance. Standards of the same copy number concentration, serially diluted from the same source material, demonstrate lower Ct values (a “shift to the left”) in the presence of F-68, indicating their higher abundance in solution due to reduced attachment to plastic. This shift translates to higher titers for AAV quantification
Fig. 10.
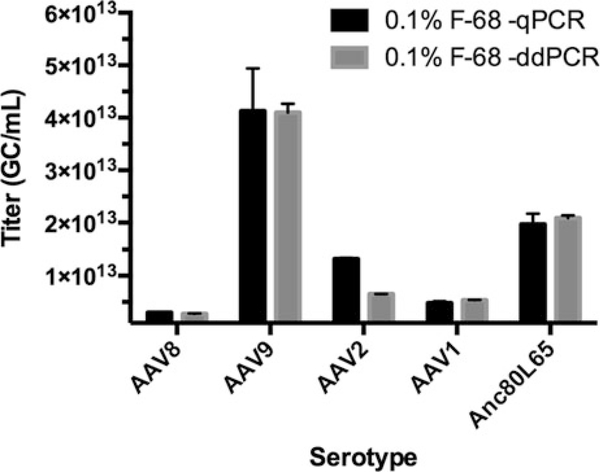
ddPCR vs. qPCR comparison across AAV serotypes carrying the same genome. The cis-plasmid used to manufacture these vectors was used for the qPCR standard. The same TaqMan™ assay was used for all measurements and exactly the same sample dilutions were assayed by both technologies. Pluronic® F-68 improves qPCR-based AAV genome quantification to a level comparable to ddPCR by making more target available for quantification and correcting qPCR standard performance
Fig. 11.
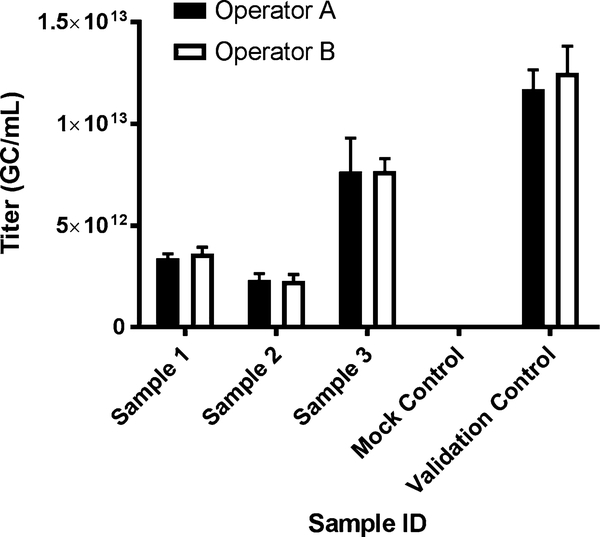
ddPCR intra-laboratory assessment of quantification variability. Trainees should first observe a run set up by a more experienced user. They should then set up their own run under supervision before taking up the assay independently. Samples of known concentration should be used to ensure accurate quantification by new users
Table 2.
Plasmid control samples for qPCR or ddPCR
| Sample ID | CMV2 Titer #1 | CMV2 Titer#2 | CMV2 Titer #3 | CMV2 Average | CMV2 Stnd Dev | Notes |
|---|---|---|---|---|---|---|
| 1E8 IE-5 (Plasmid DNase-I [−] Control) | 8.45E+07 | 8.85E+07 | 8.00E+07 | 8.43E+07 | 4.25E+06 | Copies per reaction |
| 1E8 IE 5 (Plasmid DNase-I [+] Control) | 3.25E+06 | 4.70E+06 | 5.50E+06 | 4.48E+06 | 1.14E+06 | Copies per reaction |
DNase-I efficiency is >95%. Note that contrary to observations made previously [10], we are able to quantify plasmid DNA very accurately
Acknowledgments
We wish to extend our appreciation to Qin Su and Ru Xiao, directors of Vector Production at the Horae Gene Therapy Center, UMass Medical School and the Gene Transfer Vector Core (vector.meei.harvard.edu) at the Grousbeck Gene Therapy Center, Schepens Eye Research Institute, Mass Eye and Ear, respectively, for providing valuable vectors for the comparisons presented here. This work was supported by 1P01AI100263-05, 1R01NS076991-05, R01 HL097088, and 4P01HL131471-01 (GPG) and Giving/ Grousbeck (LHV).
Footnotes
The use of in-house reagents is strongly discouraged as these are a common source of PCR contamination and variability.
SSS DNA serves as a blocking agent and nucleic acid carrier.
The microcentrifuge tubes used in this protocol are intentionally not autoclaved. This is to avoid debris or film deposits from accumulating on the internal surface of the tubes during sterilization, which may carry nucleic acid contaminants, potential PCR inhibitors, or active nucleases. Clean buffers or diluents placed in autoclaved tubes that have been heated above 37 °C may exhibit a marked increase in O.D. 260 readings compared to buffers or diluents placed in non-autoclaved tubes that are otherwise identically treated, suggesting the presence of contaminants whose identity cannot be confirmed [25].
Use of low-retention or siliconized tubes in conjunction with Plutonic® F-68 has a slight negative effect on ddPCR quantification due to an unknown interaction. Use of these treated plastics is, therefore, discouraged.
Electronic and multichannel pipettes will facilitate and speed up liquid handling, particularly for the distribution of vector diluent and sample replicates. However, they are expensive and are not an absolute requirement. AAV quantification will not be significantly affected by the use of calibrated manual pipettes. Manual LTS™ pipettes are our preferred choice due to their ergonomic piston plunging and demonstrated performance.
DNase-I is needed to digest un-encapsidated viral genomic DNA or residual plasmid DNA that may have escaped Benzonase® nuclease treatment during the recombinant AAV manufacturing process. Typical efficiency of digestion with DNase-I is 95% or greater, and should be confirmed with an appropriate plasmid control each run (Table 2).
Pluronic® F-68 solution, a.k.a. poloxamer 188 solution, is a BASF registered trademark of a 10% solution of polyoxyethylene-polyoxypropylene block copolymer, a nonionic surfactant that serves to prevent AAV attachment to plastic [20, 26]. Other surfactants or detergents may be used as well, but these will need to be properly validated. Most importantly, these reagents must not be detrimental to the function of Taq polymerase during PCR.
When F-68 is not used during AAV quantification there is a two- to tenfold drop in concentration estimates. The extent of the underestimation of titers varies according to serotype (Fig. 6). Also, in the absence of F-68, ddPCR yields higher titers than qPCR. This is most likely due to surfactants present in the droplet generation oil, the emulsification of the PCR reaction itself, or both.
The recommended final concentration of F-68 is 0.1%. We evaluated different concentrations of F-68 and found that increasing the amount of surfactant yielded variable results but did not significantly improve AAV quantification (Fig. 7). While a final F-68 concentration of 0.05% appears to be appropriate for quantification, we use 0.1% in anticipation of novel AAV variants with unique properties that may make them challenging to quantify. Diluting F-68 beyond 0.1% affects AAV serotypes differentially. The use of F-68 has consequences beyond vector production and quantification [27].
SSS DNA in the buffer will degrade over time. The sample diluent may be stored at 4 °C for no more than a week.
These solutions follow Bio-Rad’s recommended ddPCR primer and probe concentrations. The final oligo concentrations in droplets may be different than in the original 20 μL reaction, as the total volume in the reaction wells increases following droplet formation and only a fraction of this volume is composed of actual droplets. Note that if qPCR primers have already been designed for use at a lower final concentration, their Tm’s will likely increase if they are used for ddPCR at a higher concentration and the same annealing temperature. In order to keep the Tm constant at a higher oligo concentration, one would have to shorten the oligo or increase the annealing temperature. Simply increasing the concentration of qPCR primers is therefore likely to make ddPCR slightly less specific. However, we have not seen an impact on ddPCR quantification when using qPCR primers at higher concentrations. During oligo design, we generally fix the final primer, salt, and magnesium concentrations at the annealing temperature recommended by the instrument manufacturer (Bio-Rad) or set as default (Thermo Fisher), then extend or trim the oligo sequence accordingly using the Two-State Hybridization simulator of the DINAMelt application within the UNAfold Web Server [28, 29]. We seek to design stringent oligos whose Tms are not lower than 1 °C below the annealing temperature. Since the default annealing temperature for qPCR or ddPCR is 60 °C, the 7„,s of our oligos are not lower than 59 °C for specific amplification. Oligo Tms lower than 59 °C risk that the primer will not bind at the desired temperature, while much higher Tms risk off-target hybridization. The design results in an exponential overrepresentation of oligos in the reaction mixture compared to target sequences. We have found that commercial algorithms at oligo synthesis websites tend to underestimate oligo Tms when compared to the DINAMelt application. The Two-State Hybridization simulator consistently yields the shortest primers at the standard qPCR buffer composition of 50 mM Na+ or K+ and 1.5 mM Mg2+.
Primer design may be fine-tuned for specificity by performing annealing gradient PCR, but this may overlook the fact that the TaqMan™ probe will also have a large contribution to specificity. By the time the PCR cycle reaches the annealing temperature from the denaturation step, the TaqMan™ probe is already firmly bound to the target due to its 10 °C higher Tm. Other considerations for primer design apply also, depending on the complexity of the sample and the background material, which may call for more sophisticated computational resources for quality assurance. However, despite their importance, proper handling of these variables alone will not result in accurate quantification of AAV.
TaqMan™ probes are susceptible to photobleaching; therefore, shield these reagents from light as much as possible.
If unsure about the state of calibration of your instrument, check it for accuracy against a sample of known concentration such as SSS DNA, or seek professional calibration from the manufacturer. Always validate your spectrophotometer with a different instrument, different samples, and a different method of quantification. It is imperative that the spectrophotometer be properly blanked with exactly the same buffer at the same dilution used to prepare the plasmid samples that are quantified. For example, if your plasmid sample is in QIAGEN EB butter and you dilute the plasmid sample tenfold for the spectrophotometer, use a tenfold dilution of naive QIAGEN EB buffer in the same diluent to blank prior to quantification.
qPCR and ddPCR are extremely sensitive technologies [30]. Because of this, prepare the work area anew as specified between different steps in the protocol. When working with high copy number targets, aerosol formation increases the probability of cross-contamination. Contamination of negative controls is routinely encountered even with the use of brand new reagents.
The UV light should not be on longer than 15 min. The bulbs have a limited life span and a prolonged exposure will not be more effective.
It is well established that the quantification of double-stranded (ds) or self-complementary AAV (scAAV) genomes by qPCRis problematic [10, 31, 32]. While the focus of this protocol is not scAAV genome quantification, it has been suggested that a careful choice of standard should resolve most issues for scAAV quantification by qPCR [33]. This, in addition to proper standard handling, purification, and quantification, makes qPCR a competitive technology for quantification in the absence of the more costly ddPCR platform. The standard does not need to be of identical sequence and structure as the experimental vector genome in most cases, but this choice must be considered for very difficult targets.
For real-time PCR, it is important that the sequence of interest (or target) be accessible for amplification. That is, the sequence must be linear and unencumbered by secondary structures that may limit its access by Taq polymerase. Most plasmids do not need linearization, but some do. Since it is difficult to predict which plasmids will require linearization, we linearize all plasmid standards used for quantification. Supercoiled plasmids that need linearization may show a shift toward higher Ct values (less sensitivity) for the same initial copy number compared to their identical linear counterparts, and may not be detected by qPCR at all past a certain concentration. These events introduce systemic quantification errors. For TaqMan™ qPCR assay design, always check the plasmid standard sequence for secondary structures using a nucleic acid folding program at the annealing temperature [28, 29]. These technologies generate data that are highly reproducible but not necessarily accurate, and thus statistical metrics of the standards alone cannot guarantee accurate quantification. We hope to provide here a complete picture of the necessary requirements.
Do not pre-warm elution buffers. Keep all reagents, and perform all cleanup steps, at room temperature. Pre-warming elution buffers may introduce systemic quantification errors [25].
The two methods offer a cross-reference check for sample purity and accuracy of quantification. That is, for a pure species of high concentration plasmid, quantification by the two methods should significantly agree (Fig. 8). This formally links the physical measurement of the qPCR standard to the results of downstream fluorometry-based PCR methods of quantification, either qPCR or ddPCR. In the event of significant disagreement between these two methods of qPCR plasmid standard quantification, do not continue. Retrace previous steps and evaluate workflow for errors.
AAV PCR-based quantification is a sampling exercise. This means that the integrity of the sample as a unit is paramount. Whether the sample to be quantified is plasmid or virus, collect it at the bottom of the tube by centrifugation before pipetting to avoid changes in concentration due to condensation or loose sample spotting inside the tubes. Then vigorously vortex samples to ensure homogeneity. Finally, aspirate the sample by pipetting only once with proper technique. Never reintroduce the pipette tip into the sample during aspiration (or sampling) and re-aspirate, because there is a risk of enriching the pipette tip with extra material that will be detected, quantified, and increase error.
The dials of manual LTS pipettes can be set to accommodate up to three decimal points, depending on the unit. Pipette the exact intended volume by taking advantage of this feature for increased accuracy: lock the pipette dials and work within the linear range of the units when preparing standards for real-time PCR. We avoid pipetting less than 2 μL for any step.
The use of F-68 for dilution of the standard lowers the Ct values for plasmid DNA during qPCR, demonstrating that pure DNA attaches to plastic in its absence. F-68 maximizes the number of target sequences available for real-time PCR quantification (Fig. 9). The use of this excipient in the standard dilution is justified because it will also be included in all dilutions of AAV experimental samples or controls, where it will serve the same function. Lowering the Ct value of the standards by addition of F-68 will cause the experimental sample titers to decrease, but at the same time, addition of F-68 will cause more experimental sample to become available for quantification in a serotype-dependent manner. F-68, therefore, introduces a correction to quantification which predominantly results in increased titers. In real-time PCR, it is imperative that “apples-to-apples” comparisons be made. Optimal quantification is achieved with real-time PCR when standards and test samples are identical sequences and are handled within the same or nearly the same environment.
Vortexing the tube and rinsing the pipette tip are intended to keep the dilutions homogeneous during the serial dilution step. The same discipline must be applied to the dilution of AAV experimental samples or controls. We make 100 μL serial dilutions of linear standard plasmid at a time (10 μL plasmid + 90 μL AAV dilution buffer), which ensures accurate pipetting and reduced freeze-thawing cycles.
The use of a color-coded tube system is highly recommended here, as it will help organize the standards for future use, especially if there are several standards in the freezer.
The controls are as follows: DNase-I plasmid control (two reactions, with and without DNase-I), Mock Control (one reaction), and AAV Validation Control (one reaction). The no-template control (NTC) is added at the end, during the PCR plate setup. For the AAV validation control, an in-house vector from previously manufactured sample lots is sufficient. This vector can be any serotype, but its genome should carry the intended target sequence for PCR. For convenience, we recommended using a vector that includes several commonly used target sequences within a standard single-stranded recombinant AAV genome containing ITRs, promoter, transgene, and polyadenylation signal within the packaging limit of the virion. It should be a vector that may be used in an experimental setting, carrying no unusual features. Do not use an artificial composite of target sequences for PCR quantification, which may jeopardize efficient genome packaging by AAV and equal detection by PCR technologies. This control serves to measure inter-assay reproducibility and assess operator performance. We typically use a relatively high titer vector (>5E+12 GC/ mL), but this is not absolutely necessary, as quantitative PCR has a wide dynamic range and vector loss is minimal. Several such controls may be needed, depending on the particular sequence being measured.
A common source of error in these assays is inadvertent sample switching. Researchers should be vigilant at all times.
Do not add DNase-I to the reaction mix at this time. This is because one of the twro DNase-I plasmid control reactions will not include the enzyme. If more than one control set is to be included in the plate for a different TaqMan™ assay, adjust the estimation of reagent volumes accordingly.
Make sure to pick up one pipette tip at a time, and to keep the tip boxes closed whenever possible.
Keep the DNase-I cold at all times and never vigorously vortex to mix. Rather, invert the tube several times and only tap or gently pulse vortex once for mixing.
Do not vigorously vortex the PCR tube strips after adding samples or controls to the reaction mixture. DNase-I activity may be diminished.
Program the C1000 Touch thermocycler as follows: 25 °C, 2 min; 37 °C, 1 h; 4 °C, ad infinitum. We do not recommend heat-inactivating DNase-I at this time, as vector genomes will be digested if they are exposed to the enzyme after capsid breakdown, which is likely to occur at temperatures used for DNase-I inactivation. The danger is exacerbated because the temperature cools to 4 °C, briefly returning the reaction to DNase-I’s optimal temperature range after the genomes have potentially been exposed. Rather, we first dilute the DNase-I reactions (thereby diluting the enzyme), then inactivate DNase-I during PCR. DNase-I does not return to its optimal incubation temperature once thermocycling commences. We have read droplets soon after PCR, 2 h later, or the following day and have observed no significant difference in our positive control titers. Similarly, for qPCR, fluorescence is read in real-time and data collection takes place at 60 °C, well above the optimum temperature of DNase-I. This contributes to AAV titers that are equivalent between qPCR and ddPCR and highlights the importance of complementary methods of AAV quantification. Another important consideration is the length of time and the temperature at which the quantification plate is allowed to stand prior to the start of PCR. For instance, if the plate is left at room temperature for 2 h or more, the titers may significantly decrease not because AAV genomes are degraded, but because DNase-I will slowly degrade primers and probes in the reaction mixture. High-throughput qPCR may require such wait times for automated plate stacking and sequential loading, and these dangers may reduce the accuracy of titration.
After the reaction is complete, the protocol may be stopped if needed. The samples may be stored in the thermocycler overnight at 4 °C or placed on ice for transport and stored in a −20 °C freezer until the next steps can be completed. Note that the viral capsids are assumed to be intact after the DNase-I incubation. Unless otherwise noted, most AAV capsids are stable at 37 °C and the encapsidated genomes are, therefore, protected from further action by DNase-I [8, 17]. A simple check may be implemented whereby AAV samples are incubated in the presence or absence of DNase-I for quality assurance.
It is strongly recommended to use the electronic pipette that is most appropriate for the task at hand. For instance, dispensing 45 μL volumes with a 1000 μL electronic unit, although convenient, risks introducing large pipetting errors. Avoid shortcuts, if possible. Any changes must yield the same dilutions as before.
This is another stopping point. The final dilutions may be transferred to the refrigerator or freezer until the next step. Plutonic® F-68 will prevent AAV particles from adhering to the inner wall of the 0.65 mL tubes under these storage conditions [26], preserving the quantification integrity of the assay. The movement of the virions in solution will be slowed down in cold temperatures as well, with less particles reaching the inner wall of the polypropylene tube.
Next, proceed to either qPCR or ddPCR. The same dilutions may be used for either technology and inputs from both technologies will provide clues about the general quality of quantification. Successful quantification by both technologies should result in an equivalent measurement of AAV genome concentration (Fig. 10).
The steps that follow are intended for the AutoDG™ droplet generation method. If droplets are to be generated “manually,” eight reactions at a time, a manual droplet generator will be needed. Consult with Bio-Rad for details on all aspects of the droplet digital PCR system [15, 16, 34].
It may not be possible to include all controls on the plate for any one TaqMan™ assay due to space limitations. In this case, adjust the reaction map accordingly. It is imperative, however, that unassigned wells in any one column be filled with an actual (mock) ddPCR reaction, as opposed to just water or PBS, for efficient droplet generation.
A Rainin 200 μL electronic pipet will accurately deliver 18 μL of reagent, despite the fact it nominally dispenses between 20 and 200 μL accurately. Make sure this is the case before routinely implementing it in your quantification workflow.
This non-optically rated film is used to protect the reaction wells from contamination while transporting and centrifuging the plate. The film should come off easily after centrifugation.
DG32™cartridges (microfluidic channels sufficient for four columns of PCR reactions) come in a single block. If a full plate’s worth of droplets are not generated, a portion of the DG32™ cartridge may be unused (up to three columns). To prevent contamination and structural issues with the instrument, it is suggested that no attempt be made to salvage unused cartridges in a DG32™ block.
Similarly, not all tips within a tip box may be used in any one instance of droplet generation. Save the unused tips and reassemble them into full boxes inside a biosafety cabinet that has been cleaned as described in Subheading 3.1. Transfer unused tips using sterile forceps or a multichannel pipette to populate the new box of tips.
Bio-Rad recommends that reaction plates be kept at 4 °C in the thermocycler or refrigerator for at least 2 h after completion of PCR. This allows droplets to reach a proper size for subsequent reading and counting. During plate readout, as many as 85% of the droplets may be rejected in any one well. Droplet loss, however, does not appear to result in substandard quantification. Nevertheless, this recommendation should be followed, as it appears to maintain a consistent droplet count.
“Concentration” in the QuantaSoft™ .csv file refers to copies per μL in the 20 μL reaction. This number is derived from the Poisson distribution and correction that is a feature of the methodology and software, and does not yet represent concentration of the actual sample. To obtain final titers, multiply “Concentration” by 20 μL to obtain the total number of copies per reaction well, and then divide by 2 pL, the input sample volume, to obtain the concentration of the diluted sample (this is represented by the factor “10” in the formula). Multiply by the dilution factor of 1,000,000, and then multiply by 1000 to convert from GC/μL to GC/mL. For AAV experimental samples, this generally means multiplying the “Concentration” value by 1E+10 unless a different dilution factor was used during preparation. Refer to Bio-Rad’s Droplet Digital PCR Application Guide for more information [16].
This formula measures the number of plasmid copies that were added to the initial DNase-I reaction. A nominal amount of IE +08 copies in a volume of 5 μL was added. If the spectrophotometry and fluorometry measurements of the stock linear plasmid were accurate, if all subsequent serial dilutions were executed correctly, and if the surfactant F-68 performed as expected, you should obtain a value very close to 1E+08 copies. Once again, first multiply “Concentration” by 20 μL to obtain the total number of copies per reaction well. Then multiply by the dilution factor of 100,000. Finally, multiply by 2.5, as 5 μL of the linear plasmid was used in the initial DNase-I reaction, but only 2 μL of the 100,000-fold dilution was used for the ddPCR reaction. The expected number of plasmid copies in the 20 μL ddPCR reaction of the DNase-I control (without DNase-I) is 400 copies. A “Concentration” value of 20 copies/μL thus represents nearly 100% recovery of the input plasmid.
Adequate training of laboratory personnel is a necessity, irrespective of past experience. Intra-laboratory assessment of AAV quantification must be performed for protocol validation (Fig. 11).
For quality control purposes, it is strongly recommended that ddPCR results for a given TaqMan™ assay be compared against standard-based qPCR quantification during the development and validation of new assays, or for standard troubleshooting if both instruments are available. Preferably, real-time PCR validation of new assays should be carried out prior to or concomitantly with ddPCR validation runs, using the same samples to avoid introducing inadvertent biases. This will ensure that future results from one platform do not wrongly influence changes to the other. If the results do not match, revisit the entire procedure and troubleshoot as needed (Fig. 10).
All of these reactions will be run in triplicate, but for convenience we assemble one large reaction and split it into three wells of the plate using an electronic pipette. Although this is a shortcut, our experience suggests that if all reactions are set up and pipetted correctly, the outcome will be similar to performing an individual reaction for each well. For ddPCR we add any one sample or control reaction to the plate in triplicate by distributing 2 μL per well using the multi-dispense function of an electronic p20 pipette and a 6 μL aspiration. The qPCR plate is thus set up in a manner equivalent to the ddPCR plate.
In qPCR the total copies per reaction (“Qty Mean”) are provided directly from the standard curve, so no back calculation is needed as in ddPCR. To obtain GC/μL of the diluted sample, simply divide “Qty Mean” by the input volume per well: 5 μL. ssAAVs are known to package plus ( + ) and minus (−) genome strands in a 1:1 ratio [35], and the TaqMan™ probe will bind only one of these strands. Because the PCR reaction will generate a detectable AAV amplicon from an undetectable AAV genome strand, and this amplicon will amplify exponentially in subsequent cycles, there is a one cycle differential of detection between the two strands, which corresponds to a twofold reduction compared to the double-stranded linearized plasmid standard. Hence, the final titer is multiplied by 2. For scAAVs this is not needed because the two complementary AAV genome strands are combined in a single particle. Therefore, barring the issue of first-order kinetics of re-annealing, the Ct values of the standard can be compared directly with the Ct values of the scAAV genome in qPCR [33] (see Note 17).
Conflict of Interest: LHV is an inventor on gene therapy technologies licensed to various biopharmaceutical companies, a founder of Akouos and Gensight, a consultant to multiple entities with gene and genome editing therapeutic interest, and receives sponsored research from Selecta and Solid Biosciences and Lonza Houston.
References
- 1.Samulski RJ, Chang LS, Shenk T (1987) A recombinant plasmid from which an infectious adeno-associated virus genome can be excised in vitro and its use to study viral replication. J Virol 61(10):3096–3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grimm D, Pandey K, Nakai H, Storm TA, Kay MA (2006) Liver transduction with recombinant adeno-associated virus is primarily restricted by capsid serotype not vector genotype. J Virol 80(1):426M–39. 10.1128/JVL80.1.426-439.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rabinowitz JE, Rolling F, Li C, Conrath H, Xiao W, Xiao X, Samulski RJ (2002) Crosspackaging of a single adeno-associated virus (AAV) type 2 vector genome into multiple AAV serotypes enables transduction with broad specificity. J Virol 76(2):791–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hildinger M, Auricchio A, Gao G, Wang L, Chirmule N, Wilson JM (2001) Hybrid vectors based on adeno-associated virus serotypes 2 and 5 for muscle-directed gene transfer. J Virol 75(13):6199–6203. 10.1128/JVL75.13.6199-6203.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao G, Vandenberghe LH, Wilson JM (2005) New recombinant serotypes of AAV vectors. Curr Gene Ther 5(3):285–297 [DOI] [PubMed] [Google Scholar]
- 6.Clement N, Grieger JC (2016) Manufacturing of recombinant adeno-associated viral vectors for clinical trials. Mol Ther Methods Clin Dev 3:16002 10.1038/mtm.2016.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sommer JM, Smith PH, Parthasarathy S, Isaacs J, Vijay S, Kieran J, Powell SK, McClelland A, Wright JF (2003) Quantification of adeno-associated virus particles and empty capsids by optical density measurement. Mol Ther 7(1):122–128 [DOI] [PubMed] [Google Scholar]
- 8.Pacouret S, Bouzelha M, Shelke R, AndresMateos E, Xiao R, Maurer A, Mevel M, Turunen H, Barungi T, Penaud-Budloo M, Broucque F, Blouin V, Moullier P, Ayuso E, Vandenberghe LEI (2017) AAV-ID: a rapid and robust assay for batch-to-batch consistency evaluation of AAV preparations. Mol Ther 25 (6 ):1375–1386. 10.1016/j.ymthe.2017.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burnham B, Nass S, Kong E, Mattingly M, Woodcock D, Song A, Wadsworth S, Cheng SH, Scaria A, O’Riordan CR (2015) Analytical ultracentrifugation as an approach to characterize recombinant adeno-associated viral vectors. Hum Gene Ther Methods 26(6):228–242. 10.1089/hgtb.2015.048 [DOI] [PubMed] [Google Scholar]
- 10.Lock M, Alvira MR, Chen SJ, Wilson JM (2014) Absolute determination of single-stranded and self-complementary adeno-associated viral vector genome titers by droplet digital PCR. Hum Gene Ther Methods 25 (2): 115–125. 10.1089/hgtb.2013.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Werling NJ, Satkunanathan S, Thorpe R, Zhao Y (2015) Systematic comparison and validation of quantitative real-time PCR methods for the quantitation of adeno-associated viral products. Hum Gene Ther Methods 26 (3):82–92. 10.1089/hgtb.2015.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rohr UP, Wulf MA, Stahn S, Steidl U, Haas R, Kronenwett R (2002) Fast and reliable titration of recombinant adeno-associated virus type2 using quantitative real-time PCR. J Virol Methods 106(1 ):81–88 [DOI] [PubMed] [Google Scholar]
- 13.Lock M, McGorray S, Auricchio A, Ayuso E, Beecham EJ, Blouin-Tavel V, Bosch F, Bose M, Byrne BJ, Caton T, Chiorini JA, Chtarto A, Clark KR, Conlon T, Darmon C, Doria M, Douar A, Flotte TR, Francis JD, Francois A, Giacca M, Korn MT, Korytov I, Leon X, Leuchs B, Lux G, Melas C, Mizukami H, Moullier P, Muller M, Ozawa K, Philipsberg T, Poulard K, Raupp C, Riviere C, Roosendaal SD, Samulski RJ, Soltys SM, Surosky R, Tenenbaum L, Thomas DL, van Montfort B, Veres G, Wright JF, Xu Y, Zelenaia O, Zentilin L, Snyder RO (2010) Characterization of a recombinant adeno-associated virus type 2 reference standard material. Hum Gene Ther 21(10):1273–1285. 10.1089/hum.2009.223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bio-Rad (2017) Automated droplet generator instruction manual. Bio-Rad, Hercules, CA: http://www.bio-rad.com/webroot/web/pdf/lsr/literature/10043138.pdf [Google Scholar]
- 15.Bio-Rad (2017) QX200™ droplet reader and QuantaSoft™ software instruction manual, http://www.bio-rad.com/webroot/web/pdf/lsr/literature/10031906.pdf
- 16.Bio-Rad (2017) Droplet Digital™ PCR applications guide. Bio-Rad, Hercules, CA: http://www.bio-rad.om/webroot/web/pdf/lsr/literature/Bulletin_6407.pdf [Google Scholar]
- 17.Rayaprolu V, Kruse S, Kant R, Venkatakrishnan B, Movahed N, Brooke D, Lins B, Bennett A, Potter T, McKenna R, Agbandje-McKenna M, Bothner B (2013) Comparative analysis of adeno-associated virus capsid stability and dynamics. J Virol 87 (24):13150–13160. 10.1128/JVI.01415-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plumridge A, Meisburger SP, Pollack L (2017) Visualizing single-stranded nucleic acids in solution. Nucleic Acids Res 45(9):e66 10.1093/nar/gkwl297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie J, Mao Q, Tai PWL, He R, Ai J, Su Q, Zhu Y, Ma H, Li J, Gong S, Wang D, Gao Z, Li M, Zhong L, Zhou H, Gao G (2017) Short DNA hairpins compromise recombinant adeno-associated virus genome homogeneity. Mol Ther 25(6)4363–1374. 10.1016/j.ymthe.2017.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bennicelli J, Wright JF, Komaromy A, Jacobs JB, Hauck B, Zelenaia O, Mingozzi F, Hui D, Chung D, Rex TS, Wei Z, Qu G, Zhou S, Zeiss C, Arruda VR, Acland CM, Dell’Osso LF, High KA, Maguire AM, Bennett J (2008) Reversal of blindness in animal models of leber congenital amaurosis using optimized AAV2mediated gene transfer. Mol Ther 16 (3):458–465. 10.1038/sj.mt.6300389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PGR experiments. Clin Ghent 55(4):611–622. 10.1373/ciinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- 22.Huggett JF, Foy CA, Benes V, Enislie K, Garson JA, Haynes R, Hellemans J, Kubista M, Mueller RD, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT, Bustin SA (2013) The digital MIQE guidelines: minimum information for publication of quantitative digital PGR experiments. Clin Ghent 59 (6):892–902. 10.1373/clinchem.2013.206375 [DOI] [PubMed] [Google Scholar]
- 23.Wilson JM (2015) A call to arms for improved vector analytics! Hunt Gene Ther Methods 26 (1): 1–2. https://doi.org/l0.1089/hgtb.2015.1502 [DOI] [PubMed] [Google Scholar]
- 24.Mueller C, Ratner D, Zhong L, EstevesSena M, Gao G (2012) Production and discovery of novel recombinant adeno-associated viral vectors. Curr Protoc Microbiol Chapter 14:Unitl4D 11 10.1002/9780471729259.ntcl4d01s26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis LK, Robson M, Vecherkina Y, Ji C, Beall G (2010) Interference with spectrophotometric analysis of nucleic acids and proteins by leaching of chemicals front plastic tubes. BioTechniques 48(4):297–302. 10.2144/000113387 [DOI] [PubMed] [Google Scholar]
- 26.Torcello-Gontez A, Wulff-Perez M, GalvezRuiz MJ, Martin-Rodriguez A, CabrerizoVilchez M, Maldonado-Valderranta J (2014) Block copolymers at interfaces: interactions with physiological media. Adv Colloid Interface Sci 206:414–27. 10.1016/j.cis.2013.10.027 [DOI] [PubMed] [Google Scholar]
- 27.Mason JB, Gurda BL, Van Wettere A, Engiles JB, Wilson JM, Richardson DW (2017) Delivery and evaluation of recombinant adeno-associated viral vectors in the equine distal extremity for the treatment oflanrinitis. Equine Vet J 49(1 ):79–86. 10.1111/evj.12547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Markham NR, Zuker M (2005) DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res 33(Web Server issue): W577–W5 81. https://doi.org/l0.1093/nar/gki591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Markham NR, Zuker M (2008) UNAFold: software for nucleic acid folding and hybridization. Methods Mol Biol 453:3–31. 10.1007/978-1-60327-429-6-1 [DOI] [PubMed] [Google Scholar]
- 30.Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, Bright IJ, Lucero MY, Hiddessen AL, Legler TC, Kitano TK, Hodel MR, Petersen JF, Wyatt PW, Steenblock ER, Shah PH, Bousse LJ, Troup CB, Mellen JC, Wittmann DK, Erndt NG, Cauley TH, Koehler RT, So AP, Dube S, Rose KA, Montesclaros L, Wang S, Stumbo DP, Hodges SP, Romine S, Milanovich FP, White HE, Regan JF, Karlin-Neumann GA, Hindson CM, Saxonov S, Colston BW (2011) High-throughput droplet digital PGR system for absolute quantitation of DNA copy number. Anal Chem 83(22):8604–8610. 10.1021/ac202028g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fagone P, Wright JF, Nathwani AC, Nienhuis AW, Davidofif AM, Gray JT (2012) Systemic errors in quantitative polymerase chain reaction titration of self-complementary adeno-associated viral vectors and improved alternative methods. Hum Gene Ther Methods 23 (1): 1–7. 10.1089/hgtb.2011.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Ling C, Song L, Wang L, Aslanidi GV, Tan M, Ling C, Srivastava A (2012) Limitations of encapsidation of recombinant self-complementary adeno-associated viral genomes in different serotype capsids and their quantitation. Hum Gene Ther Methods 23 (4):225–233. 10.1089/hgtb.2012.090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wagner A, Rohrs V, Kedzierski R, Fechner H, Kurreck J (2013) A novel method for the quantification of adeno-associated virus vectors for RNA interference applications using quantitative polymerase chain reaction and purified genomic adeno-associated virus DNA as a standard. Hum Gene Ther Methods 24 (6): 3 55–363. 10.1089/hgtb.2013.095 [DOI] [PubMed] [Google Scholar]
- 34.Bio-Rad (2017) QX200™ Droplet Generator Instruction Manual. Bio-Rad, Hercules, CA: http://www.bio-rad.com/webroot/web/pdf/lsr/literature/10031907.pdf [Google Scholar]
- 35.Rose JA, Berns KI, Hoggan MD, Koczot FJ (1969) Evidence for a single-stranded adenovirus-associated virus genome: formation of a DNA density hybrid on release of viral DNA. Proc Nad Acad Sci USA 64(3):863–869 [DOI] [PMC free article] [PubMed] [Google Scholar]