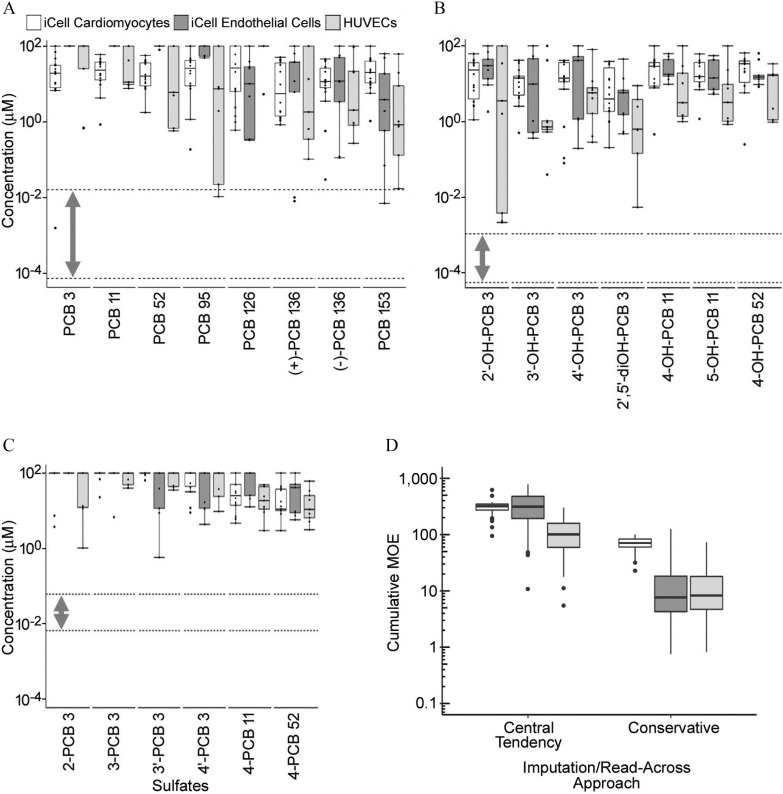
Figure 7.
Characterization of the margins of exposure for (A) PCBs, (B) OH-PCBs, and (C) PCB sulfates separately, as well as for the cumulative effect all serum PCBs and metabolites together using an imputation/read-across approach (D). In (A–C), box plots show distributions of point-of-departure values (one standard deviation of the variability in cells treated with vehicle) for various phenotypes sorted by cell type (white: iCell®-cardiomyocytes; dark gray: iCell®-endothelial cells; light gray: human umbilical vein ECs (HUVECs). Dotted lines with bidirectional arrows are indicative of human serum concentration ranges for selected PCBs, OHPCBs, and PCB sulfates. In (D), box plots show the distribution across individuals in the biomonitoring studies (Ampleman et al. 2015; Grimm et al. 2017; Koh et al. 2016a, 2016b), of their cumulative margin of exposure across all PCBs and metabolites. Each cell type is cumulated separately, and both a “central tendency” and a “conservative” imputation/read across approach are used. All box plots were generated in ggplot2 R package (version 3.1; R Development Core Team) and indicate 25th, 50th (median), and 75th percentiles. Whiskers indicate the smallest and largest values within interquartile range above the 75th or below the 25th percentile. Outliers are indicated as dots. Chemical names and abbreviations of PCBs and metabolites are listed in Table S1. Note: CMs, cardiomyocytes; ECs, endothelial cells; HUVECs, human umbilical vein endothelial cells; OHPCBs, hydroxylated metabolites of PCBs; PCBs, polychlorinated biphenyls.