Fig. 1. Wildlings resemble wild mice and differ significantly from conventional laboratory mice in their bacterial microbiome at major microbial niches and immunological barrier sites.
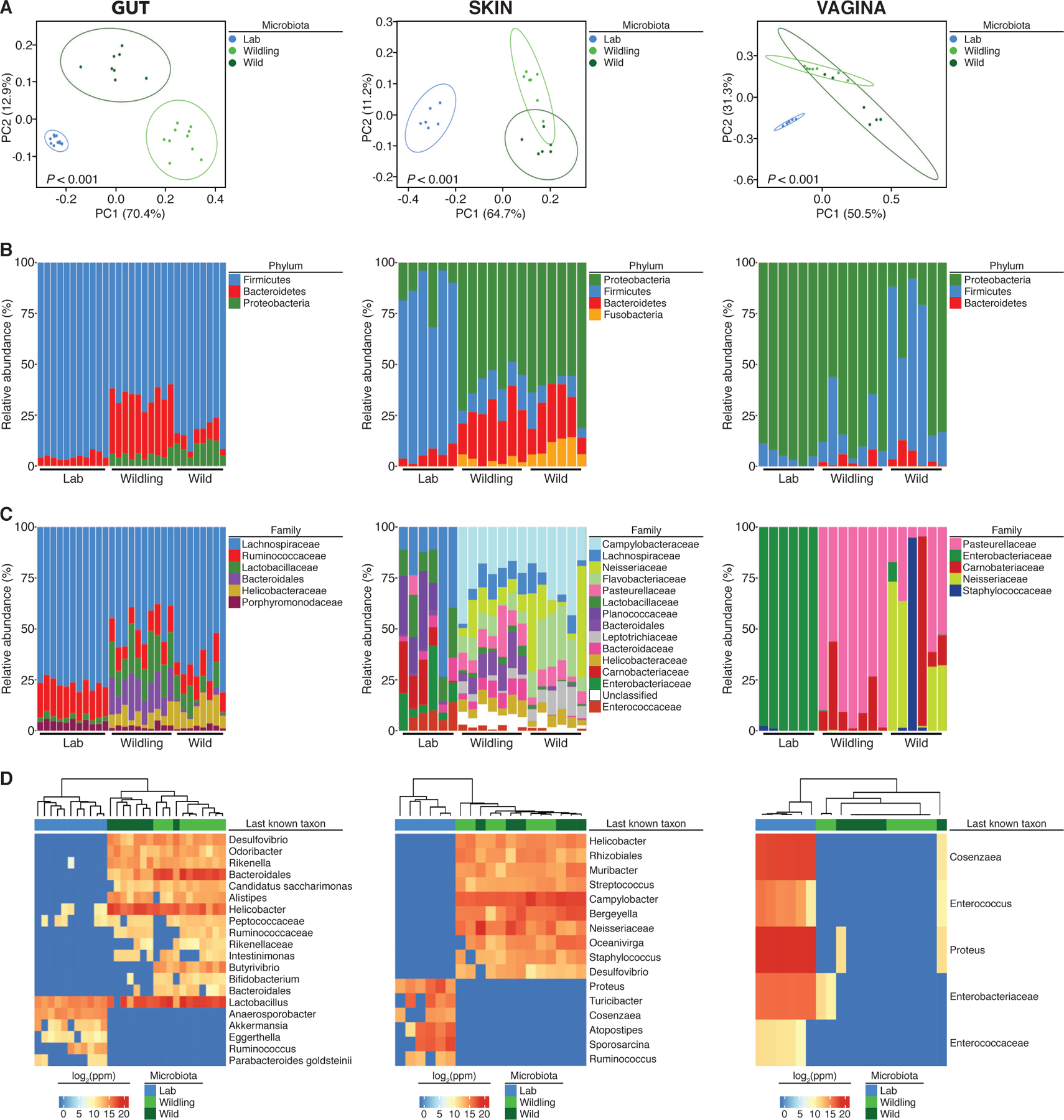
The bacterial microbiome of wildlings, wild mice (Wild), and conventional laboratory mice (Lab) was profiled at gut, skin and vagina by 16S rRNA sequencing: (A) Weighted UniFrac PCoA. (B and C) Relative abundance at the rank of phylum and family. (D) The heatmap shows the last known taxa with greatest variance among groups (log2-fold change values). Data shown are from 6 to 11 independent biological replicates per group. Each skin and vaginal replicate represents tissue pooled from 3 mice. Significance in (A) was determined by PERMANOVA.
