Fig. 3.
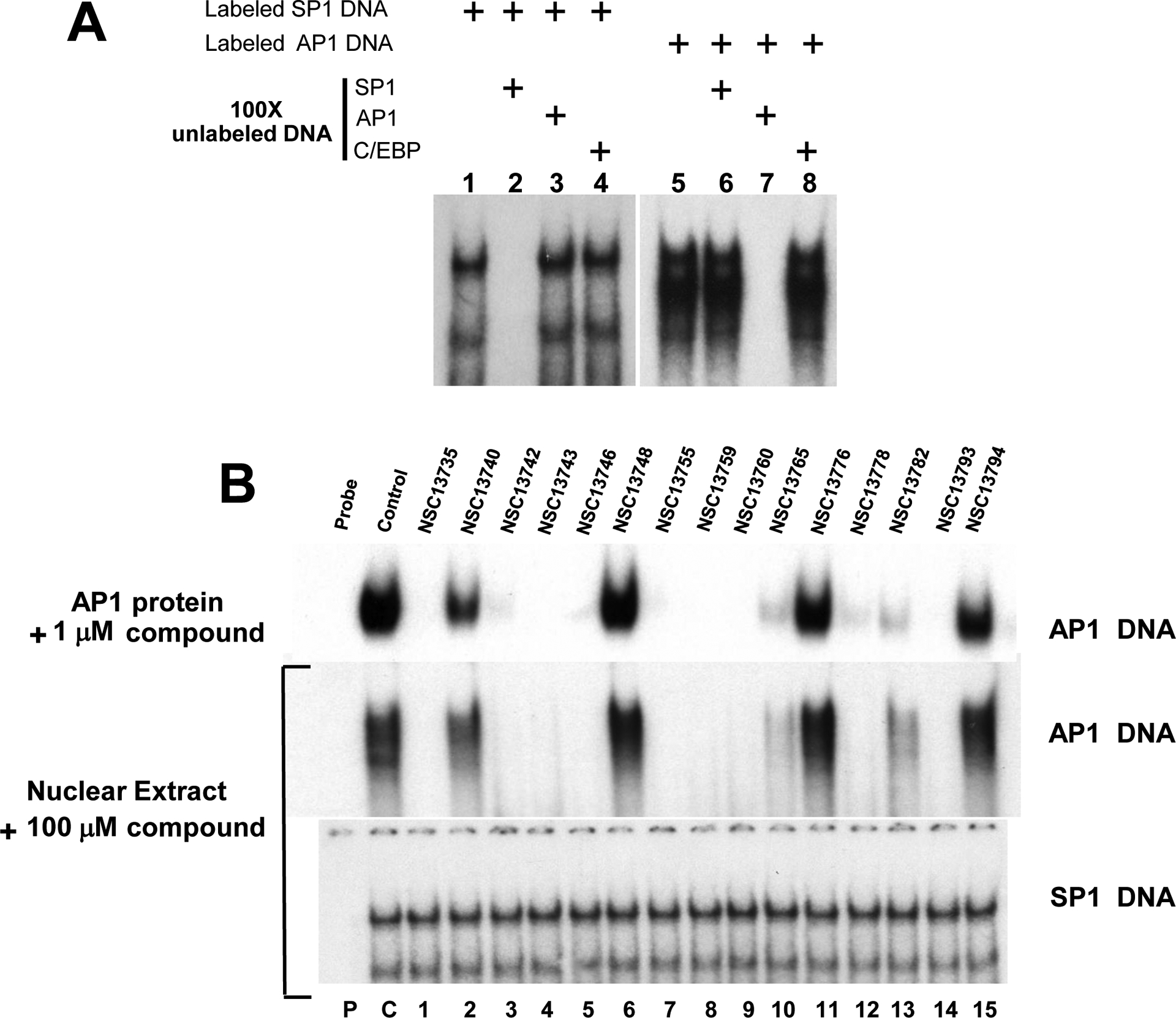
EMSA using nuclear extracts and CD studies show the inhibiting activities and specificity of compound(s). A) Competition assay using AP1 and SP1 probe. Lanes 1–4 and 5–8 represent binding results using mouse liver nuclear extracts mixed with AP1 and SP1 probe respectively. SP1 binding is totally abrogated by 100 fold excess of unlabeled SP1 probe only whereas excess amount of unlabeled AP1 and C/EBP probes have no effect. Similarly AP1 activity (lanes 5–8) can be inhibited only by excess unlabeled AP1 probe (lane 7). These results show that SP1 and AP1 proteins bind specifically to their corresponding probe in nuclear extracts. B) EMSA using mouse nuclear extracts and AP1 probe produces similar results as pure B-ZIP protein. Upper panel shows the DNA binding of pure c-Fos|JunD (AP1) B-ZIP domains to an AP1 containing oligonucleotide in the presence of 1 μM compounds (Fig. 2B). Bottom two panels show EMSA using mouse liver nuclear extracts showing the DNA binding activity to either a consensus AP1 or SP1 DNA binding site in the presence of antimony compounds. P, C and numbers 1–15 have the same meaning as in Fig. 2.
