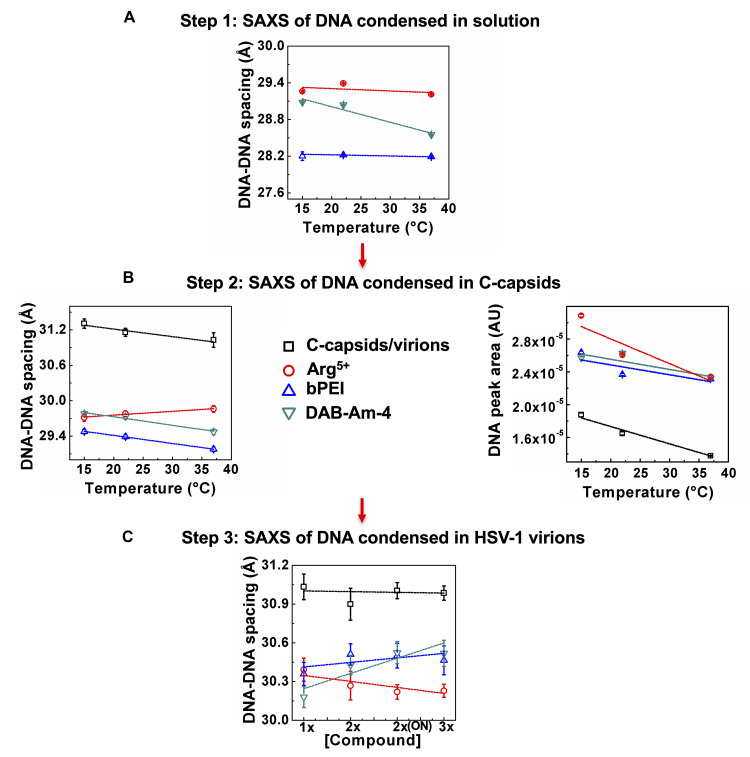
Fig 2.
(A) Addition of DAB-Am-4, bPEI 600, or Arg5+ compounds condenses free dsDNA in solution, with resulting DNA-DNA interaxial distances, ds, below 31Å (this value is measured for DNA packaged in HSV-1 capsid without compound addition at physiologic conditions). ds -values were collected at 15, 23, and 37°C and showed minor variation with temperature. (B and C) SAXS screening assay showing that the three selected DNA-condensing compounds mentioned in (A) can penetrate the HSV-1 C-capsid (B) as well as the lipid-membrane-enveloped virions (C) and condense the packaged DNA inside, reducing DNA-DNA interaxial spacing, ds, below 31Å. We tested two- and three-fold higher compound concentration (2x and 3x) than the minimum required for DNA condensation at the N/P ratio of 1.5 used in our study. Incubation time with the compound was 30 min or 12 h (ON: overnight). Temperature and the compound incubation time had only a small effect on DNA-DNA spacing, suggesting that the compound capsid and lipid envelope permeability kinetics are not limited by the diffusion rate. Vertical error bars are from the non-linear fitting of the DNA diffraction peak with a Gaussian function with background subtraction.