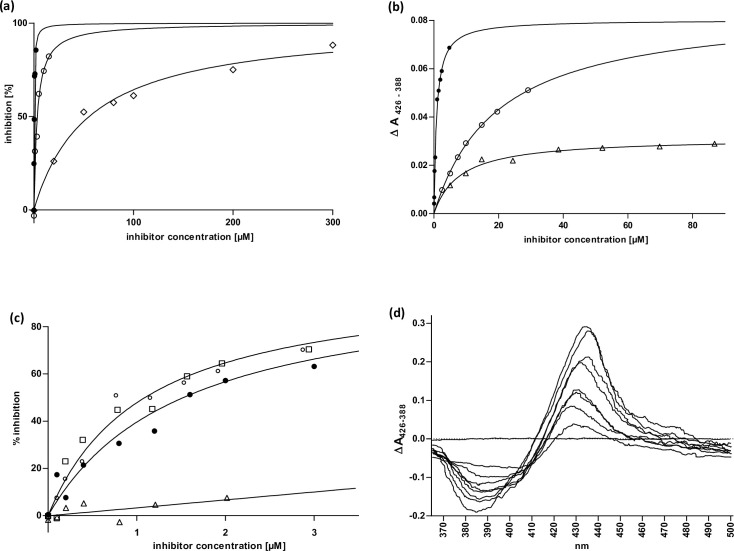
Fig 1. Enzyme kinetics and ligand binding in subcellular systems (microsomes + supersomes).
(a) CYP2E1 inhibition by 12-imidazolyl-dodecane derivatives. Varying concentrations of the inhibitors 12-Imidazolyl-dodecanol (I-ol) (●), 1-Imidazolyl-dodecane (I-an) (○) and 12-Imidazolyl-dodecan-1-yl-phosphocholine (I-phosphocholine) (◇) were added to human CYP2E1 reconstituted system to calculate Ki values. Nonlinear fitting was achieved using competitive inhibition as model including an offset in the case of I-phosphocholine (b) KD of imidazolyl-dodecane derivatives. Difference spectra of rat liver microsomes titrated with the inhibitors I-ol (●), I-an (○) and I-phosphocholine (△) were recorded limited by the solubility of the reagents. Experimental data were fitted to an equation including one specific binding site. (c) CYP2E1 inhibition by ω-imidazolyl-alkyl derivatives. Varying concentrations of the inhibitors 7-Imidazolyl-heptanol (△), 9-Imidazolyl-nonanol (□), 10-Imidazolyl-decanol (○) and I-ol (●) were added to a human CYP2E1 reconstituted system in supersomes to calculate Ki values. Nonlinear fitting was achieved using competitive inhibition as model. (d) Difference spectra of 12-Imidazolyl-dodecan-1-yl-phosphocholine. It showed a characteristic Soret peak at 425–435 nm and minimum at 390–410 nm, which corresponds to a typical type II difference spectrum and is induced by substances both providing a bond to the trivalent heme iron (III) and interacting with the active center of the enzyme.