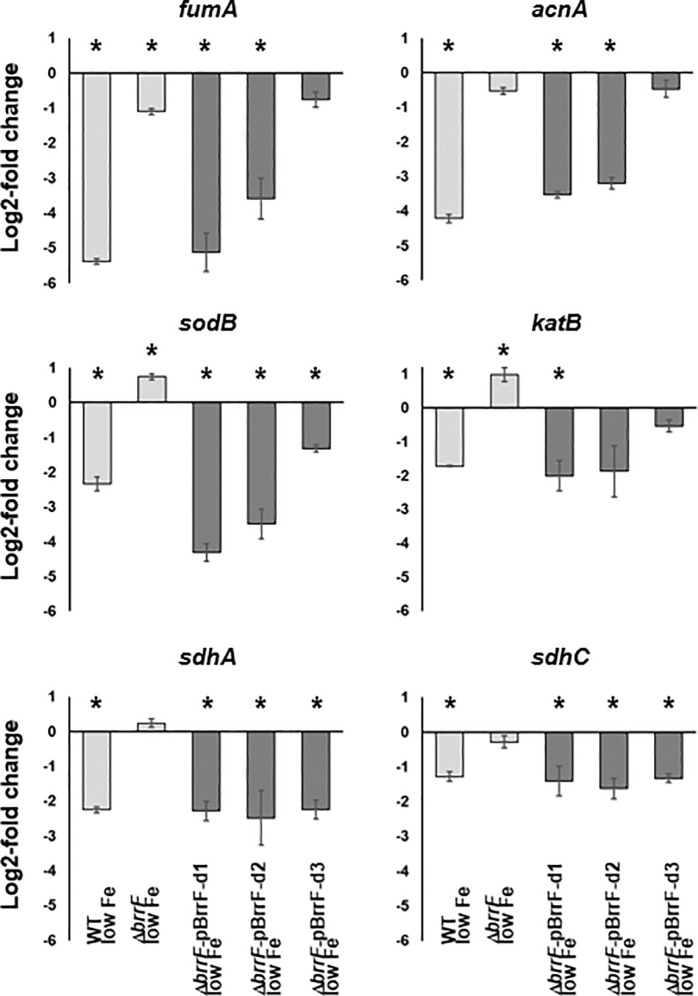
Fig 5. BrrF-dependent expression of computationally predicted targets under low iron condition.
Cells were harvested for RNA extraction after “pulse-expression” of 30 min in a mid-log culture. Downregulation of gene expression could be complemented in ΔbrrF by overexpressing BrrF in trans. Complementation was dependent on the 5’end of BrrF for most genes except for sdhA and sdhC. Light grey: Expression compared to iron-replete condition. Dark grey: Expression compared to ΔbrrF-vector control. pBrrF-d1 and pBrrF-d2: plasmids overexpressing native BrrF with or without processing. pBrrF-d3: derivative of pBrrF-d2 with point mutations near the BrrF 5’end. Asterisks: Significant difference to the respective control, with p ≤ 0.05.