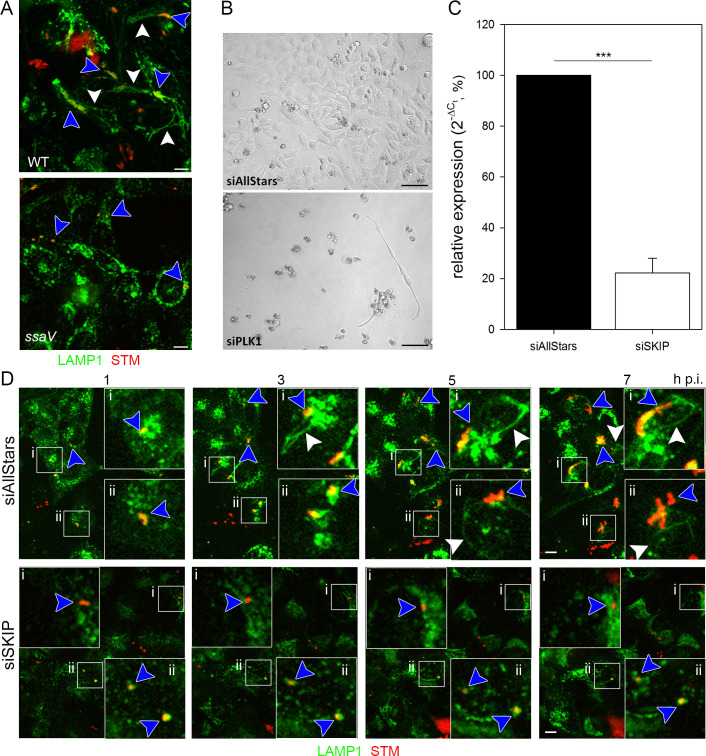
Fig 1. RNAi screen setup and validation.
A) Intracellular phenotypes of STM under screening conditions. HeLa LAMP1-GFP cells were infected with mCherry-labelled STM WT or ssaV strains and imaged live 8 h p.i. by SDCM. Images display the presence of STM in LAMP1-positive SCV (blue arrowhead), the induction of SIF formation by STM WT (white arrowhead), and the lack of SIF formation by the STM ssaV strain. Scale bar, 10 μm. B) Controls for siRNA-mediated knockdown. HeLa LAMP1-GFP cells were reverse transfected with scrambled AllStars siRNA or PLK1 siRNA for 72 h and then imaged. Scale bar, 20 μm. C) Validation of SKIP siRNA knockdown. HeLa LAMP1-GFP cells were reverse transfected with AllStars or SKIP siRNA for 72 h. Then, RT-PCR targeting SKIP was performed. Depicted is the mean with standard deviation of three biological replicates (n = 3) each performed in triplicates. Statistical analysis was performed using Student’s t-test and indicated as: ***, p < 0.001. D) SKIP knockdown as a control for the inhibition of SIF formation. HeLa LAMP1-GFP cells were first reverse transfected with AllStars or SKIP siRNA for 72 h. Then, cells were infected with mCherry-labelled STM WT (MOI = 15) and imaged live by SDCM 1–7 h p.i in hourly intervals. Blue arrowheads indicate SIF-forming or non-SIF-forming single bacteria or microcolonies, white arrowheads indicate SIF. Scale bar, 10 μm.