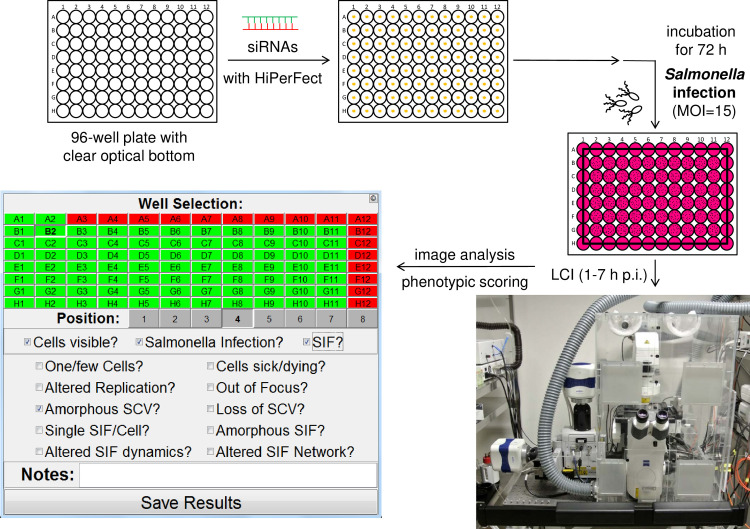
Fig 2. Basic workflow of the trafficome RNAi screen.
24 96-well plates with clear optical bottoms per biological replicate (with 3 biological replicates in total, n = 3) were automatically spotted with siRNAs. HeLa LAMP1-GFP cells were seeded for reverse transfection with each plate also containing negative, positive, and phenotype-specific controls. After 72 h of incubation, infection with STM WT (and ssaV as control, MOI = 15) was performed, followed by LCI for the acquisition of eight positions per well with single bacteria-focused Z-planes with hourly intervals of imaging from 1–7 h p.i. on an SDCM system. Subsequent phenotypic scoring was performed using the SifScreen utility. The MATLAB-based data input mask allows the entry of well- and position-specific information on general cell behavior and Salmonella/SMM phenotypes and the generation of a results report.