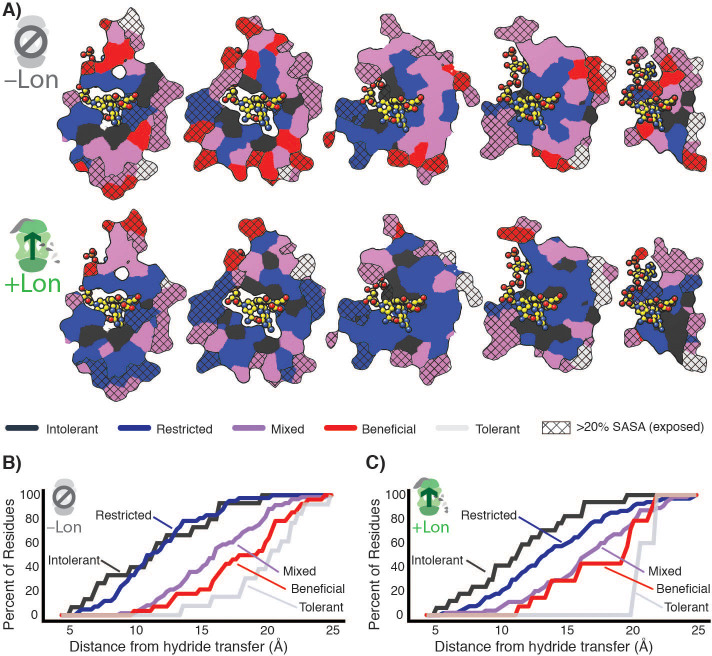
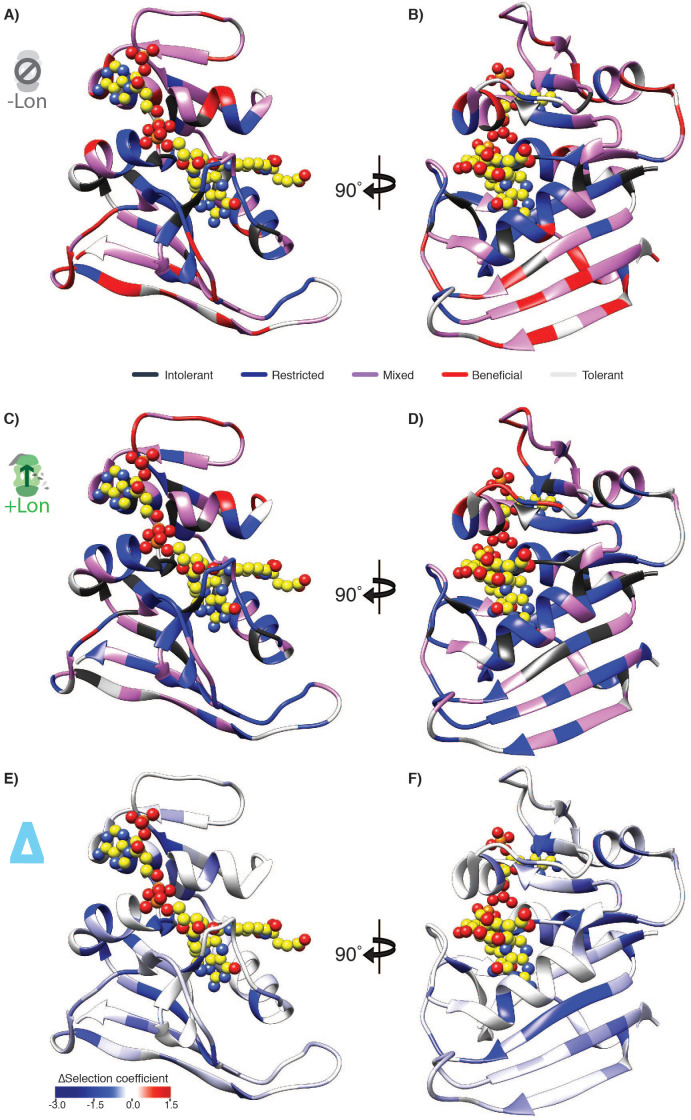
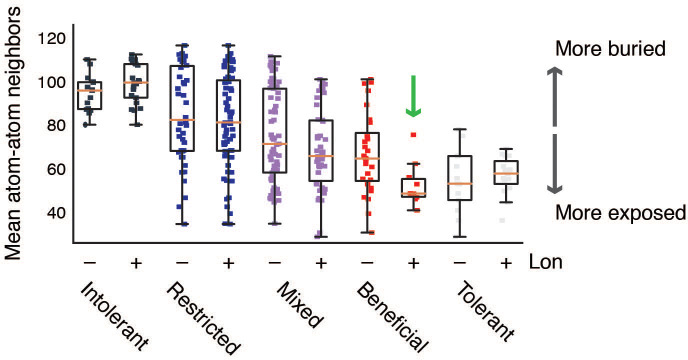
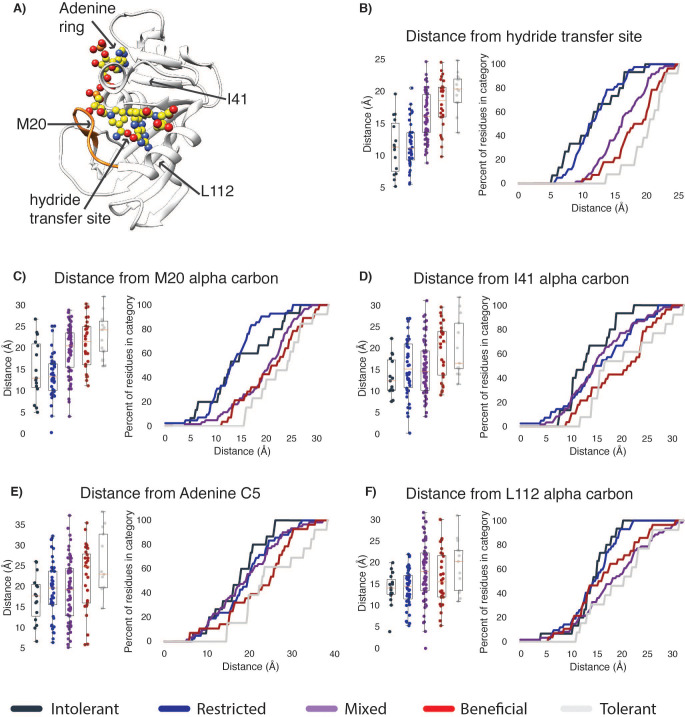
Figure 5. Structural characterization of multiple constraints on the DHFR mutational landscape.
(A) Mutational response categories from –Lon selection (top, categories in Figure 2C,D) and +Lon selection (bottom, categories as in Figure 2C,D) colored onto residues and displayed on slices as in Figure 1E. (B) Relationship between mutational response and distance from hydride transfer for –Lon selection. The percent of positions from each mutational response category are plotted as a function of distance from the site of hydride transfer. Each category colored as in A), top). (C) Relationship between mutational response and distance from hydride transfer for +Lon selection. Each category colored as in A), bottom).