Figure 1.
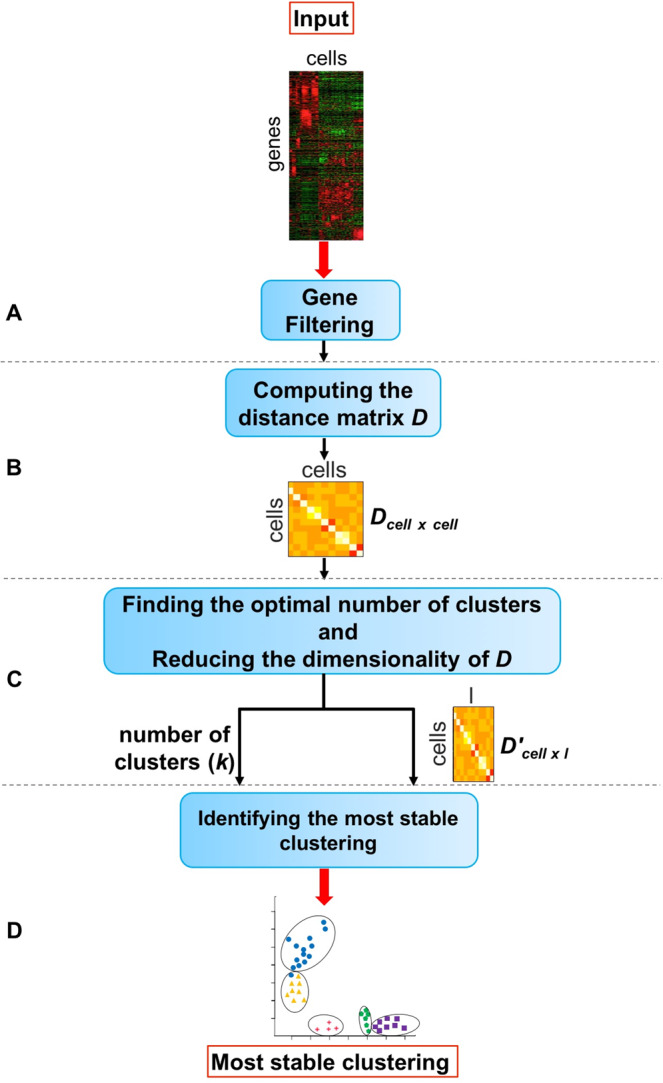
The overall workflow of the proposed method. Given the single cell gene expression matrix, module (A) eliminates the genes that are not expressed in any cell. Using the resulting matrix, module (B) computes the Euclidean distance between the cells. The output of this module is a distance matrix in which the rows and columns are the cells (Dcell×cell). Module (C) reduces the dimensionality of the distance matrix using the t-distributed stochastic neighbor embedding (t-SNE) technique. In this module, an average silhouette method is employed to choose the optimal number of clusters k. Finally in module (D), the lower-dimension distance matrix and the optimal number of clusters k obtained from module (C) are used as the input data to identify the most stable clustering of cells. Figure 2 shows the details of module D.
