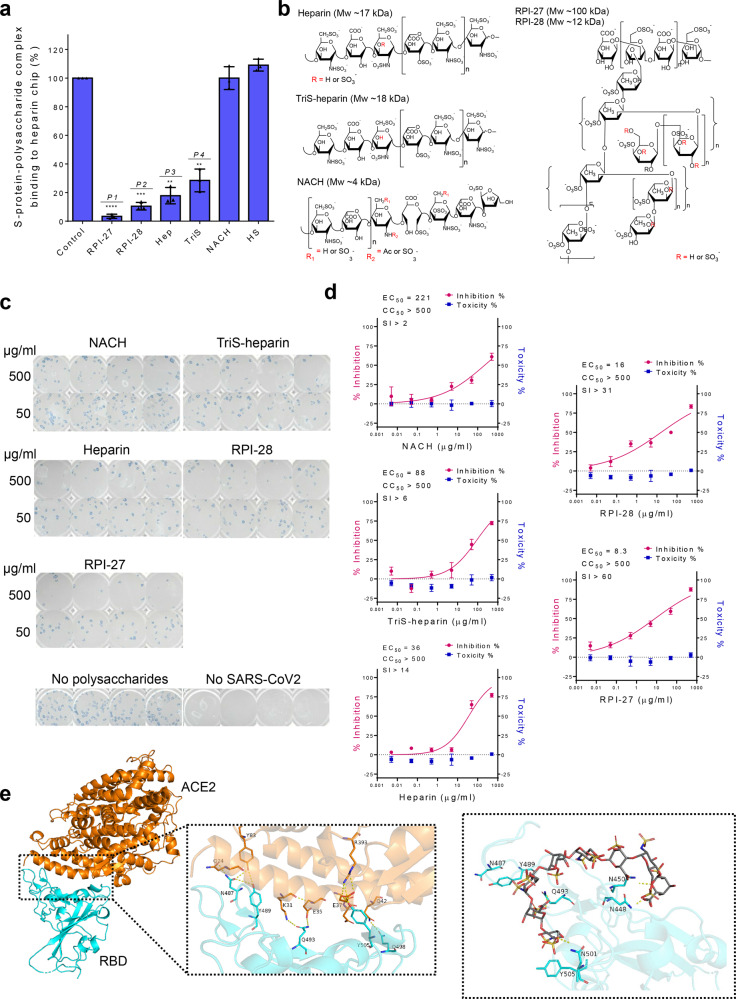
Fig. 1. Assessment of antiviral activities of certain sulfated polysaccharides.
a Surface plasmon resonance (SPR) experiments were used to screen polysaccharides that outcompete immobilized heparin binding to SARS-CoV-2 S-protein. Data are presented as mean±s.d., n=3 biologically independent samples. A two-sided t-test was performed to test significance against the control (P1<0.0001, P2=0.0003, P3=0.0016, P4=0.0041). b Structural units comprising polysaccharides used for in vitro antiviral studies. c Focus reduction assay images of virus infection on treatment of indicated polysaccharides. At 48h after infection, Vero cells were fixed and probed with SARS-CoV-2 spike primary antibody (1:10000, Sino Bio Inc.) and HRP-conjugated goat rabbit (1:10000, Abcam) secondary antibody. d Vero cells were infected with SARS-CoV-2 at a MOI of 2.5×10−3 at different doses of each polysaccharide for 48h. The viral yield was quantified using a focus reduction assay. Cytotoxicity in Vero cells was measured using a WST-1 assay. The left and right y-axis of the graphs represent mean % inhibition of virus yield and cytotoxicity of the polysaccharides, respectively. Cytotoxicity experiments were performed in duplicate with n=3 biologically independent samples. Focus reduction assay experiments were performed in mean±s.d. (quadruplicate measurements) with n=3 biologically independent samples. e The RBD-ACE2-binding interface is stabilized by an extensive hydrogen bonding network involving sidechains of several residues on both RBD and ACE2. Polar sidechains of N487, Y489, Q493, Q498, and Y505 on the spike protein RBD along with other residues would be able to bind to heparin and inhibit RBD-ACE2 interaction. Heparin (here an octasaccharide) forms a hydrogen bond network with N448, N450, Q493, and N501 that aids in its occupancy of this binding regions and sterically restrict access to Q498, Y489, and Y505 necessary for ACE2 receptor binding.