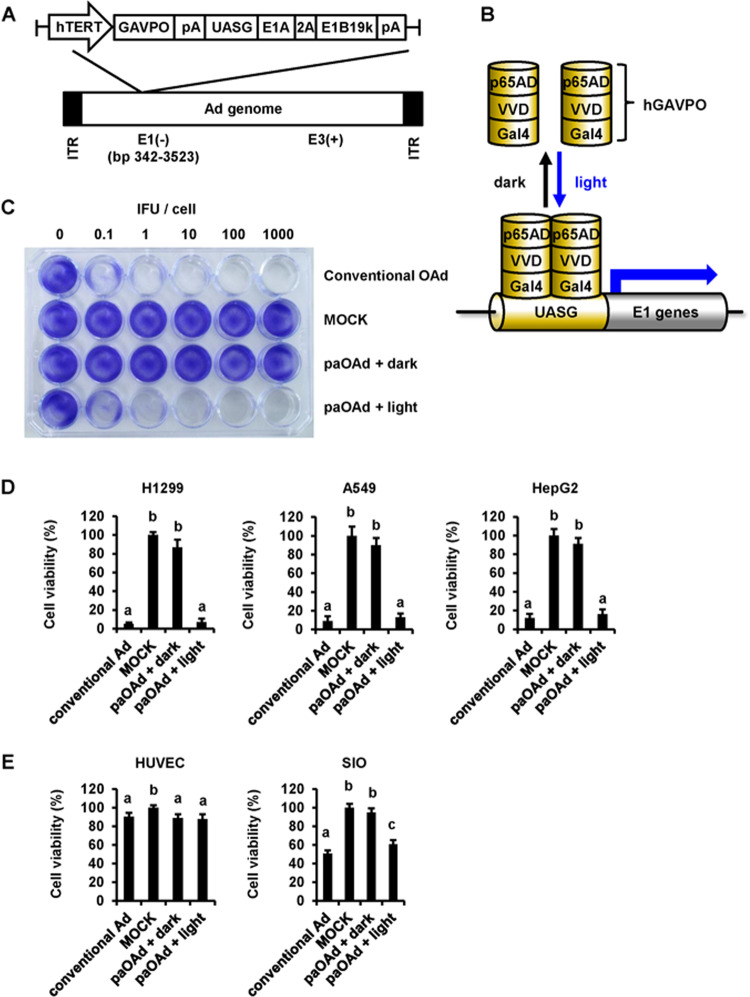
Fig. 1. Design and characterization of the photoactivatable oncolytic adenovirus.
a Construction of the photoactivatable oncolytic adenovirus (paOAd). The E1A and E1B19 kDa (E1B19k) genes, which are necessary for adenoviral replication, are inserted downstream of the upstream activating sequence of Gal (UASG). The GAVPO construct consists of sequences encoding VVD (the smallest LOV domain-containing protein), Gal4 (Gal4 residues 1–65), and the p65 activation domain. GAVPO is under the control of the hTERT promoter. The phTERT-GAVPO-pA-UAS-E1A-2A-E1B19k-pA sequence was inserted into the E1 region-deleted human adenovirus type 5 (Ad5) backbone. b Schematic overview of the paOAd. Blue light irradiation induces homodimerization of GAVPO, which enables Gal4(65) to bind UASG, thereby inducing the expression of E1A and E1B19k. c Crystal violet analysis of the cytopathic effects of paOAd in H1299 tumor cells. The H1299 cells were infected with conventional OAd or paOAd at the indicated infectious units (IFU)/cell for 2 h followed by 0.1 mW/cm2 blue light irradiation for 5 days. Five days after infection, the cells were stained with crystal violet. Data are representative of three independent experiments. d H1299, A549, and HepG2 cancer cell lines were infected with the paOAd at 10 IFU/cell for 2 h followed by 0.1 mW/cm2 blue light irradiation for 5 days. Five days after infection, the cell viability was determined by WST-8 assay. e The normal human cells, HUVEC (TERT negative), and human SIO (TERT positive) were infected with the paOAd at 10 IFU/cell for 2 h followed by 0.1 mW/cm2 blue light irradiation for 5 days. Five days after infection, the cell viability was determined by WST-8 assay. The data were normalized by the data of the mock-infected group. The results are shown as the mean ± S.E. (n = 3). Statistical significance was evaluated by one-way ANOVA followed by Tukey’s post hoc tests. Groups that do not share the same letter had cell viabilities significantly different from each other (p < 0.05).