Abstract
Background
Adverse drug reactions (ADRs) are one of the leading causes of morbidity and mortality in health care. Understanding which drug targets are linked to ADRs can lead to the development of safer medicines.
Methods
Here, we analyse in vitro secondary pharmacology of common (off) targets for 2134 marketed drugs. To associate these drugs with human ADRs, we utilized FDA Adverse Event Reports and developed random forest models that predict ADR occurrences from in vitro pharmacological profiles.
Findings
By evaluating Gini importance scores of model features, we identify 221 target-ADR associations, which co-occur in PubMed abstracts to a greater extent than expected by chance. Amongst these are established relations, such as the association of in vitro hERG binding with cardiac arrhythmias, which further validate our machine learning approach. Evidence on bile acid metabolism supports our identification of associations between the Bile Salt Export Pump and renal, thyroid, lipid metabolism, respiratory tract and central nervous system disorders. Unexpectedly, our model suggests PDE3 is associated with 40 ADRs.
Interpretation
These associations provide a comprehensive resource to support drug development and human biology studies.
Funding
This study was not supported by any formal funding bodies.
Keywords: Adverse drug reactions, Adverse event report, FAERS, Secondary pharmacology, Machine learning, statistical modeling, Drug discovery & development, Drug safety
Research in Context.
Evidence before this study
Adverse reactions of marketed and approved drugs cause a significant amount of morbidity and mortality. Previous studies have mostly focused on identification of adverse drug reactions from post-marketing adverse event reports in patients. However, drugs can often bind many different protein targets and it remains unclear which of these targets cause adverse effects in the human body. A better understanding of the links between target engagement and the manifestation of adverse effects in patients may offer an alternative yet still mechanistically-grounded approach to predict and improve drug safety. Such in silico drug screening algorithms have thus far been limited in scope.
Added value of this study
In this work, we have leveraged adverse drug reaction events from post-marketing identification surveys and target-based in vitro pharmacology of over 2000 marketed drugs. Through machine learning, we can systematically predict the drug effects on human patient populations from their target-based preclinical profiles. We validate our machine learning predictions extensively based on chronological event reporting, comparison with drug labels and through systematic text mining of scientific literature. Through our target-centric approach, we identify 221 statistical associations between protein targets and adverse reactions, which provide novel insight into the molecular components underlying physiological adverse reactions. Our combined analysis of these two large datasets thus provides a significant advance in the field of drug safety prediction. Furthermore, these machine learning algorithms are scalable and adaptable to similar datasets, and can be accessed for download online.
Implication of all the available evidence
Taken together, we envisage that our target - adverse drug reactions associations and predictive model may accelerate drug discovery and development efforts as well as inform future human biology studies. We posit that our findings have the potential to mitigate drug safety risks already at the preclinical stage. This could lead to faster and more accurate identification of safe therapeutic candidates.
Alt-text: Unlabelled box
1. Introduction
Toxicity is one of the major causes of termination, withdrawal, or labeling of a drug candidate or drug, other than lack of efficacy [1], [2], [3]. There is an urgent need to better identify toxic on- and off-target effects on vital organ systems especially for cardiovascular, renal, hepatic and central nervous system (CNS)-related toxicities; furthermore, there is a desire to reduce cost and labor in preclinical assays and drug testing on non-human species [4], [5], [6]. In vitro pharmacological assays have been widely used to screen for possible off-targets and potential adverse effects and eliminate compounds that are not safe enough in the drug development stage as early as possible [5,7]. However, systematic prediction of compound safety and potential adverse events associated with a compound is still a challenge for the pharmaceutical industry.
Machine learning has been shown to be insightful for many different stages of drug discovery and development [4,[8], [9], [10], [11], [12], [13], [14], [15]], such as preclinical pharmacology [4], clinical trials [16], and basic science research [13,15]. Previous studies have predicted efficacy [15], target binding [4] or absorption, distribution, metabolism, and excretion (ADME) properties [17] of small molecules based on their chemical structure. However, the diversity of structures that interact with targets, even when they are well described like human Ether-a-go-go-related gene (hERG), make it challenging to produce reliable models [18]. Several studies provide small, hand-curated databases providing up to 70 pharmacological targets (i.e. receptors, ion channels, transporters, etc.) with established links to adverse side effects based on a scientific literature search [5,7,[19], [20], [21]]. Natural language processing of scientific literature [22,23] and drug labels [24] as well as databases, such as the US Food and Drug Administration (FDA) Adverse Event Reporting System (FAERS) [25], OMOP [26] and EU-ADR [27], further provide resources for machine learning approaches to learn associations between drugs and adverse drug reactions (ADRs) [4,[8], [9], [10], [11],22,28,29]. FAERS is a voluntary, post-marketing pharmacovigilance tool that can be used to monitor the clinical and post-marketing performance of drugs. Another study highlights the importance of predicting the likelihood of clinical trial side effects using human genetic studies of drug-targeted proteins [16]. From a pharmacogenomics perspective, predicting drug-target interactions using pharmacological similarities of drugs and FAERS can be beneficial for drug repositioning and repurposing [30].
In this study, we explore an alternative use of FAERS data to predict compound safety using Medical Dictionary for Regulatory Activities (MedDRAⓇ [31]) terms, which we envision to be useful for future preclinical studies. Our machine learning approach is different from the aforementioned approaches (Supplementary Table 1) because we not only predict adverse drug reaction occurrences of drugs but most importantly also extract biologically meaningful target-ADR links. Using an in vitro secondary pharmacology dataset of more than 2000 marketed or withdrawn drugs (see Methods), we built a random forest model with the subset of 1329 drugs that had accompanying adverse event reports from FAERS, to predict drug-ADR and target-ADR associations. We validate drug-ADR predictions through systematic Side Effect Resource (SIDER [24]) drug label analysis and 221 target-ADR predictions through systematic literature co-occurrence analysis. Furthermore, we find canonical target-ADR associations, such as hERG binding causing cardiac arrhythmias. We also encountered unexpected associations which warrant further investigations, such as a link between Phosphodiesterase 3 (PDE3) and several ADRs, including congenital renal and urinary tract disorders. We conclude our study with potential targets that are associated with cardiovascular and renal ADRs to demonstrate the utility and possible impact of this method in drug development and preclinical safety sciences by enabling prediction of ADRs from in vitro pharmacological profiles.
2. Methods
2.1. In vitro secondary pharmacology assays for marketed drugs
AC50 values of 2134 marketed drugs (Supplementary Table 2) were measured in up to 218 different in vitro secondary pharmacology assays. Compounds were obtained from the Novartis Institutes of Biomedical Research (NIBR) compound library and tested in a panel of in vitro biochemical and cell-based assays at Eurofins and at NIBR in concentration-response (8 concentrations, half-log dilutions starting at 30 µM). Assay formats varied from radioligand binding to isolated protein to cellular assays. Example protocols may be found at https://www.eurofinsdiscoveryservices.com/cms/cms-content/services/in-vitro-assays/. Normalized concentration response curves were fitted using a four parameter logistic equation with internally developed software (Helios). The equation used is for a one site sigmoidal dose response curve Y as a function of tested concentrations X: Y(X)=A+(B-A)/(1+(X/C)D), with fitted parameters A=min(Y), B=max(Y), C=AC50 and exponent D. By default, A is fixed at 0, whereas B is not fixed.
If a drug was not tested against a specific assay, the AC50 value was set to NA (not available). AC50 values from similar assays with the same gene target were merged to reduce the NA data and features in the random forest model; this procedure resulted in 184 different target assays (Supplementary Table 3). In case any merged assays had multiple AC50 values for the same drug, we averaged these geometrically to take into account variation over orders of magnitudes. The drugs are classified according to their annotated Anatomical Therapeutic Chemical (ATC) code [33]. In case of multiple ATC codes, we assigned the most frequent level 1 code.
2.2. Mining adverse event reports of marketed drugs using openFDA
In this study, we utilized openFDA to acquire FAERS reports related to the query compounds [25,34]. This Elasticsearch-based API provides raw download access to a large volume of structured datasets, including adverse events reports from FAERS.
We used generic compound names (e.g. “Amoxicillin”) to query through the openFDA interface, accessed programmatically using Python. In order to maximize the coverage over FDA datasets, we normalized generic names to uppercase format followed by a name similarity metric to filter out unrelated records in our analysis. We included reports when the Jaro similarity between the query generic name and reported compound name was equal or greater than 0.8. To illustrate, to query “3alpha-Androstanediol”, we acquired reports including “3ɑ-Androstanediol”, “Androstanediol”, “3-alpha-Androstanediol” as different lexical variations of the generic name and collated the resulting adverse event reports.
As the FAERS database contains information voluntarily submitted by healthcare professionals, consumers, lawyers and manufacturers, adverse event reports may be duplicated by multiple parties per event, and may be more likely to contain incorrect information if submitted by a non-medical professional. To reduce reporting bias and increase report information accuracy, we only analysed reports submitted by physicians (data field: ‘qualification’ = 1). In this subset of adverse event reports, the data were further filtered by reported drug characterization, which indicates how the physician characterized the role of the drug in the patient's adverse event. A drug can be characterized as a primary suspect drug, holding a primary role in the cause of the adverse event (data field: ‘drugcharacterization’ = 1); a concomitant drug (‘drugcharacterization’ = 2); or an interacting drug (‘drugcharacterization’ = 3). Here, we included only primary suspect drug reports. Without this restriction, model performances did not improve. We obtained all adverse events reports corresponding to the query compound that passed through the aforementioned filters.
Adverse event report descriptions are coded as medical terms of MedDRA terminology [31]. Medical observations can be reported using 5 hierarchical levels of medical terminology, ranging from a very general System Organ Class term (e.g. gastrointestinal disorders) to a very specific Lowest Level Term (e.g. feeling queasy). Each term is linked to only one term on a higher level. For each report, we recorded all MedDRA Reaction terms (data field: “reactionmeddrapt”) at the Preferred Term level and mapped these Preferred Terms to Higher Level Group Term and System Organ Class level. For each (ADR term, drug) tuple, we then calculated the ADR occurrence, defined as the following fraction: number of adverse event reports containing that ADR term relative to the total number of ADR reports for that drug.
For different FAERS versions (Q4_2014, Q4_2018 and Q2_2019), we used the same query except the time parameter TO, which was set to 12/30/2014 for the Q4_2014 query. For the other two queries, we did not set the limit parameter which was filled with the query time by default (query date was 10/10/2018 for Q4_2018 and 08/12/2019 for Q2_2019).
2.3. Random forest models and statistical methods of drug - ADR associations
To construct and train our random forest models, we used AC50 values for a panel of target assays for marketed drugs (model input; independent variable) and ADR occurrences of the compounds (model output/predictions; dependent variable). Since there may be several ADRs associated with any given drug, we took a “first-order strategy”, i.e. we assume there is no correlation between different ADRs, and a “divide and conquer” strategy, i.e. we decompose our learning task into n independent binary classification problems, where n is the number of different ADR terms in our output data (n = 26 for SOC and n = 321 for HLGT level respectively). We built a random forest [69] binary classifier for each ADR using Binary Relevance with the random forest modeling option in mldr package [70] and utiml package in R [71].
To define the features for the random forest models, we discretized and one-hot encoded our input AC50 values. Discretization was essential to limit the number of features and enhance the predictive power of the model. We defined 3 classes (levels) of AC50 ranges for each target assay (reported in Supplementary Table 2 with level values 2, 1, and 0, respectively).
-
•
Highly active class: AC50 in [0, 3 μM]
-
•
Active class: AC50 in [3 μM, 30 μM]
-
•
Inactive class: AC50 greater than 30 μM
If the AC50 value is NA, the values for all Classes are 0. Each drug has AC50 values for 184 (merged) assays, so there are 184×3 = 552 binary features to represent our input data. Features consisting of only 0 values were removed, resulting in 413 input features used for model construction.
The observed ADR occurrences retrieved from FAERS were discretized into binary dependent variables through a statistical model based on the binomial distribution (binomial model). Intuitively, a drug has an association with the ADR if the occurrence is higher than through random reporting. To formalize this, first let Nd be the total number of ADR reports for a given drug. The probability to observe an ADR occurrence OADR = X / Nd at random is equivalent to choosing that ADR X times out of Nd with X distributed binomially: X∼bin(Nd, p = 1/n). Here, n represents the total number of ADRs as defined above. Under this null distribution, we calculate the p-values for all observed ADR occurrences OADR for a given drug, and then perform a Benjamini-Hochberg False Discovery Rate (FDR) correction (using the Python statsmodels package). If an FDR-corrected p-value is < 0.01, then the ADR value for that drug is 1, reflecting an association; 0 otherwise. With Empirical-Bayes Regression-adjusted Arithmetic Mean (ERAM) [8,[36], [37], [38]], ADRs at the HLGT level were considered an association with a drug if 1.5 < ER05 < ER95 and no association otherwise [8,[36], [37], [38]]. These ERAM binary drug-ADR predictions were then compared to those from our binomial model using a χ2-test.
All random forest models were first trained using 5-fold cross validation and each fold is selected sequentially. The training set consists of 1329 drugs, which has at least one ADR report. 1063 drugs were used for training and 266 drugs were used for testing in each fold, and the distribution of drug classes (ATC) in our training set (1329 drugs) is preserved in 5-fold cross validation splits (Supplementary Fig. 1a), i.e. the drug classes are represented in 5-fold cross validation splits the way they are represented in our training set. For a given (drug) input of AC50 values and ADR, the random forest model output, termed ADR probability, can be interpreted as the probability that the ADR is associated with the drug. To enable direct comparison of model predictions with binarized ADR occurrences, we binarized these ADR probabilities with a simple threshold value of 0.5. These binary values were used for training, cross validation and to calculate classification performance metrics. All models have been constructed the same way regardless of different FAERS versions.
We evaluated our models based on six metrics: accuracy, Matthew's correlation coefficient (MCC), precision, recall, area under the receiver operating characteristic curve (AUROC) and area under the precision recall curve (AUPRC). These metrics are calculated using their definitions below, except 3 metrics: (1) MCC, which is calculated using mltools package in R [https://github.com/ben519/mltools], (2) AUROC, which is calculated using the precrec package in R [72] and (3) AUPRC, which is calculated using the PRROC package in R [73].
-
•
Accuracy =
-
•
Precision =
-
•
Recall =
-
•
MCC =
-
•
AUROC
-
•
AUPRC =
where true positive rate (TPR) is by definition equal to the recall defined above and FPR (false positive rate = ). The corresponding metrics for each individual ADR model are calculated as above. The global model statistics were obtained through calculation over all ADR models. For the X and Y randomization models, we first randomly permuted the feature rows (X randomization) or binomial model drug-ADR associations binary class values that were used for model training (Y randomization) and subsequently trained, 5x cross validated and calculated 5x CV performance metrics as described above.
2.4. Statistical methods of target-ADR associations
To find associations between gene target assays and ADRs, we first generated ADR probabilities specific to a given assay. As a model input, one out of its three random forest input features’ value was set to 1 and all others to 0. This simulates the scenario of an in silico compound that is potent with an AC50 value in the range corresponding to the positive feature only. We then utilized the ADR's random forest model, pre-trained on all available marketed drug data (see previous section), to calculate the resulting ADR probability. We repeated this procedure for each feature of all assays and each ADR.
To select the predictive features for a given ADR, we ordered the pre-trained random forest model's input features according to their Gini importance score [74] and denote the top 5% as significant features. Our criteria for a gene (target assay) - ADR pair were:
-
•
For a given ADR: at least 2 out of 3 assay features need to be significant in order to make a reliable comparison between the ADR probabilities with respect to AC50 values.
-
•
At least one of the ADR probabilities of the significant features has to be larger than zero.
We filtered out target-ADR pairs if the ADR term maps to the following SOC classes, which are not specific to body parts or underlying human biology:
-
•
general disorders and administration site conditions
-
•
injury, poisoning and procedural complications
-
•
investigations
-
•
neoplasms benign, malignant and unspecified [incl cysts and polyps]
-
•
poisoning and procedural complications
-
•
social circumstances
-
•
surgical and medical procedures
To ensure the reproducibility of the target-ADR pair selection procedure, we repeated the random forest model training with different seeds for a total of 5 times. We then took the union of the 5 sets of target-ADR pairs and discarded pairs that were only found once out of 5 runs. Finally, to determine if the mean ADR probabilities between the selected AC50 classes were statistically significantly different, we performed a two-sample t-test with sample sizes equal to the number of times a class was selected (ranging from 2 to 5 times) using the Python scikit.stats package. In case all three AC50 classes were represented, we tested the highly active versus inactive class. We then performed a Benjamini-Hochberg FDR correction. If the FDR-corrected p-value is < 0.1, then the target-ADR pair is considered a statistically significant association.
To evaluate the relation between the HLGT level ADR term hepatic and hepatobiliary disorders and target assay BSEP, we also trained and analysed two random forest models as described above to find target-ADR pairs but with only the BSEP assay data discretized with class boundaries [0, 30 μM), [30, 300 μM] and >300 μM or [0, 100 μM), [100, 300 μM] and >300 μM.
2.5. Side effect resource (SIDER) analysis
The Side Effect Resource (SIDER; version 4.1) was downloaded (http://sideeffects.embl.de/download/; accessed on 09/16/2019). The file meddra_all_se.tsv.gz contains drug-ADR pairs extracted from drug labels using text mining [24]. The supplied MedDRA preferred term (PT) was mapped to HLGT used for the random forest modeling. The file drug_atc.txt provides mappings from drug names as used in SIDER to Anatomical Therapeutic Chemical (ATC) codes. ATC codes for the 805 drugs in the test set were obtained from the NIBR compound database, and matched to ATC codes from SIDER. For drugs that could not be matched via ATC codes, additional matches were obtained by mapping the compound name, first trying the name in its entirety (e.g. “butriptyline hydrochloride”, then on the first word in the drug name (e.g. “butriptyline”). All matches, whether obtained on ATC codes or by drug name, were reviewed manually for accuracy.
2.6. Systematic validation of predicted target-ADR association using PubMed database
We built a query based on 254 unique HLGT level ADR terms and 106 unique target genes (corresponding to the assays), for which we could find a corresponding MeSH term, to retrieve linked publication identifiers (PMIDs) from the PubMed database. All PMIDs were acquired by submitting a query for every MeSH entity separately via the PubMed API engine, a search engine that provides access to the MEDLINE database of references and abstracts on life sciences and biomedical articles. Next, we determined the PMIDs for a gene-ADR pair as the intersection of the two PMID sets of each corresponding MeSH term query. Furthermore, for each possible gene-ADR pair we determined whether it was part of the 221 predicted associations from the Random Forest model or not. In this way, we obtained 219 unique positive gene-ADR pairs and a total 26,705 unique negative pairs. Lastly, we generated a set of negative pairs corresponding to all permutation pairs from the 39 unique genes and 131 unique ADRs that are part of the positive set, resulting in 4890 unique negative pairs in this negative control set. To assess any statistical overrepresentation, we calculated the number of pairs with at least one co-occurrence publication for both negative and positive sets and assessed significance with a Fisher Exact test (Python function scipy.stats.fisher_exact). Furthermore, we calculated the co-occurrence “lift” over the reporting probability when assuming independence, defined as with the total number of PMIDs in the PubMed database in 2019 (https://www.nlm.nih.gov/bsd/licensee/2019_stats/2019_LO.html). and are respectively the number of retrieved PMIDs for a unique gene-ADR pair, ADR, or gene target separately. To assess the location differences of the above described positive versus negative distribution of lift values, we performed a Mann Whitney U test (Python function scipy.stats.mannwhitneyu, two-sided, continuity correction=True).
2.7. OMOP benchmark comparison analysis
The OMOP benchmark consists of 399 drug-ADR pairs with a binary ground truth (association or no association) for 4 different ADRs at the Preferred Term (PT) level [26]. For our benchmarking, we mapped these PT ADRs to their HLGT parent term to enable comparison with our model predictions. ERAM and binomial model binary drug-ADR association predictions (both dependent on the availability of FAERS adverse event reports) were generated as described above. RF Train and RF Test indicate the performance of the RF ADR models, after training as described above, using the in vitro profiles of drugs from the overlap of drugs present in OMOP with the drugs that were also present in RF training (n = 1329 drugs) and test (n = 805 drugs) sets, respectively. Performance metrics were calculated as described above.
3. Results
3.1. Systematic in vitro pharmacology of marketed and withdrawn drugs
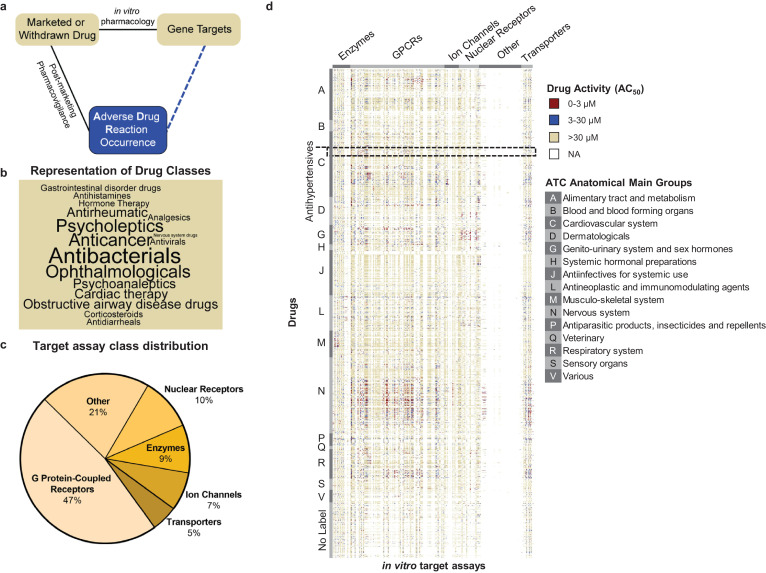
To link gene targets to ADR occurrence, we utilized in vitro pharmacology assay data for 2134 marketed or withdrawn drugs, generated by Novartis, and ADR reports from FAERS (Fig. 1a, Supplementary Table 2). Withdrawn drugs and their assay data are also included due to the fact that they are associated with a plethora of ADRs, and thereby constitute an important resource for our predictive approach. Fig. 1b summarizes the top 50% of frequently occurring primary indications, classified by the Anatomical Therapeutic Chemical (ATC) codes, of the 2134 drugs using a word cloud. The categories that have the highest number of compounds are antibacterial, ophthalmological, and antineoplastic drugs. The in vitro pharmacology assay data includes AC50 values for each drug at up to 218 different assays for 184 gene targets (see Supplementary Table 3). There are 6 classes of these 184 gene targets, with the majority [47%] of targets falling into G protein-coupled receptors (GPCRs) (Fig. 1c), which is a dominant, widely studied drug target family, broadly represented by marketed drugs [32]. Fig. 1d is a heatmap visualization of the in vitro pharmacology assay data, where each row is a drug, grouped by their ATC anatomical main group terms [33]; each column is a target assay, grouped by target class; and each value is the AC50 of drugs for target assays. Even though 70% of drug-assay combinations have not been tested, i.e. these combinations have NA value for AC50, our data indicate relatively uniform assaying with respect to the different drug classes.
Fig. 1.
Major elements of the target-ADR association analysis.
a. Schematic outline of target-ADR pair determinations. The observed relations (solid lines) between drugs and adverse drug reactions (ADRs) are determined by post-marketing pharmacovigilance and between drugs and their (off) targets by in vitro pharmacology. This approach enables prediction of associations (dashed line) between targets and ADRs through random forest modellng.
b. Representation of drug classes in word cloud. The cloud displays the top 50% most frequently occurring drug classes, representing 2134 drugs, in the Novartis in vitro pharmacology data warehouse. Size of the font of the drug class reflects the number of associated drugs.
c. Target class distribution in the Novartis in vitro secondary pharmacology assay panel. The 184 targets in the Novartis assay panel cover 6 target classes. Almost half of the target assays belong to the G protein-coupled receptor (GPCR) class.
d. Novartis target panel potency (AC50) heatmap. The profile consists of the AC50 values of 184 target assays for 2134 drugs. We considered an AC50 value less than 3 μM as highly active (red), between 3 μM and 30 μM as active (blue), and greater than 30 μM as inactive (yellow). No data for a drug-target pair is labeled as NA (white). Drugs are grouped (vertically) by their Anatomical Therapeutic Chemical (ATC) codes. Assays are grouped (horizontally) by target class.
3.2. Analysis of adverse event reports from FAERS connects drugs with human ADRs
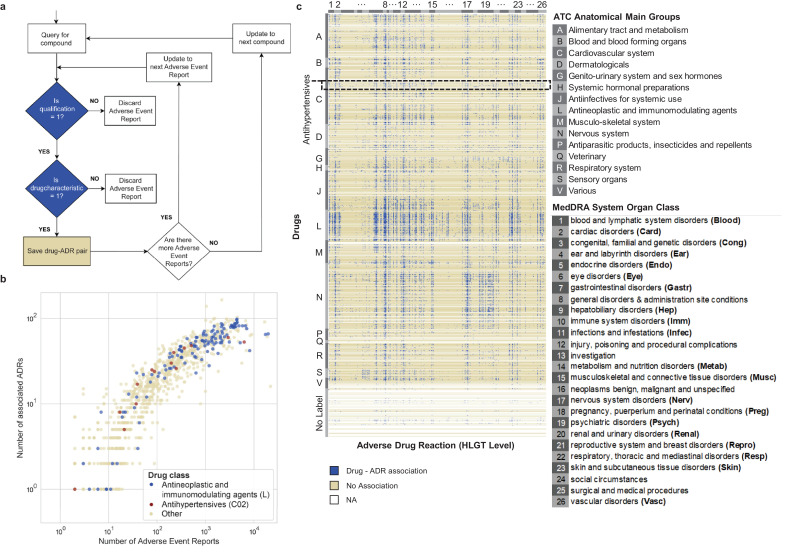
We queried FAERS [25] using openFDA [34] for 2134 marketed or withdrawn drugs in October 2018 (FAERS Q4_2018 version; covering all reports from January 2004 to October 2018) and retrieved 671,143 adverse event reports using our data extraction criteria (Fig. 2a). We only included reports which were submitted by physicians and were annotated as the primary suspect drug [35]. There are 464 drugs that did not have a matching name in FAERS, 341 drugs that did not have any adverse event reports, and 1329 drugs with at least 1 adverse event report. Intuitively, a drug is associated with a particular ADR if its reporting occurrence is higher than reports with randomly selected ADRs. To formalize this, we developed a significance test based on a binomial null distribution and false discovery rate (FDR) multiple testing correction to determine if the observed ADR occurrence was significantly high to be classified as an association (or alternatively no association) between ADR and drug (see Methods for detail). The resulting drug-ADR associations corresponded strongly (odds ratio = 11, χ2-test, p-value < 10–16) with those identified with ERAM (Empirical-Bayes Regression-adjusted Arithmetic Mean), an established Bayesian method based on the proportional reporting ratio adjusted for covariates (age group, sex and reporting year) and concomitant drugs [8,[36], [37], [38]]. Overall, we observe a positive trend between the number of adverse event reports and the number of ADR associations (Fig. 2b). Antineoplastic and immunomodulatory drugs (Fig. 2b, blue, N = 155) have many ADR associations while the extent of ADR association for antihypertensive drugs (Fig. 2b, red, N = 35) varies more widely. As an example, we visualized our drug-ADR associations (Fig. 2c), in which ADRs are grouped by MedDRA System Organ Class (SOC) level terms and drugs are grouped by ATC anatomical main group terms [33], revealing that ADRs are widespread across organs caused by antineoplastic and immunomodulating agents (Fig. 2c, label L), as well as nervous system drugs (Fig. 2c, label N).
Fig. 2.
Retrieval of Adverse Event Reports from the FDA Adverse Event Reporting System (FAERS) database.
a. Flow chart of the programmatic strategy for Adverse Event Report retrieval from FAERS by using openFDA. ‘is qualification = 1′ is a positive filter for adverse event reports that were reported by physicians. ‘is drugcharacterization’ = 1 is a positive filter for drugs that are annotated as the primary suspect drug, which hold a primary role in the cause of the adverse event.
b. Scatter plot of the number of associated ADRs for drugs as a function of the number of adverse event reports retrieved for each drug (Ndrugs = 1329). Drugs without any reported ADR are not shown.
c. Heatmap of ADR profiles (discretized as used for input of random forest model) for all marketed drugs used in this study (Ndrugs = 2134). Drugs are clustered (vertically) according to their ATC drug classes (A-V, or No label if without any ATC code) and HLGT (high level group term) ADRs are grouped (horizontally) according to the parent System Organ Class (SOC) level listed in the legend.
3.3. Random forest model learns relationship between in vitro pharmacology and reported ADRs in humans
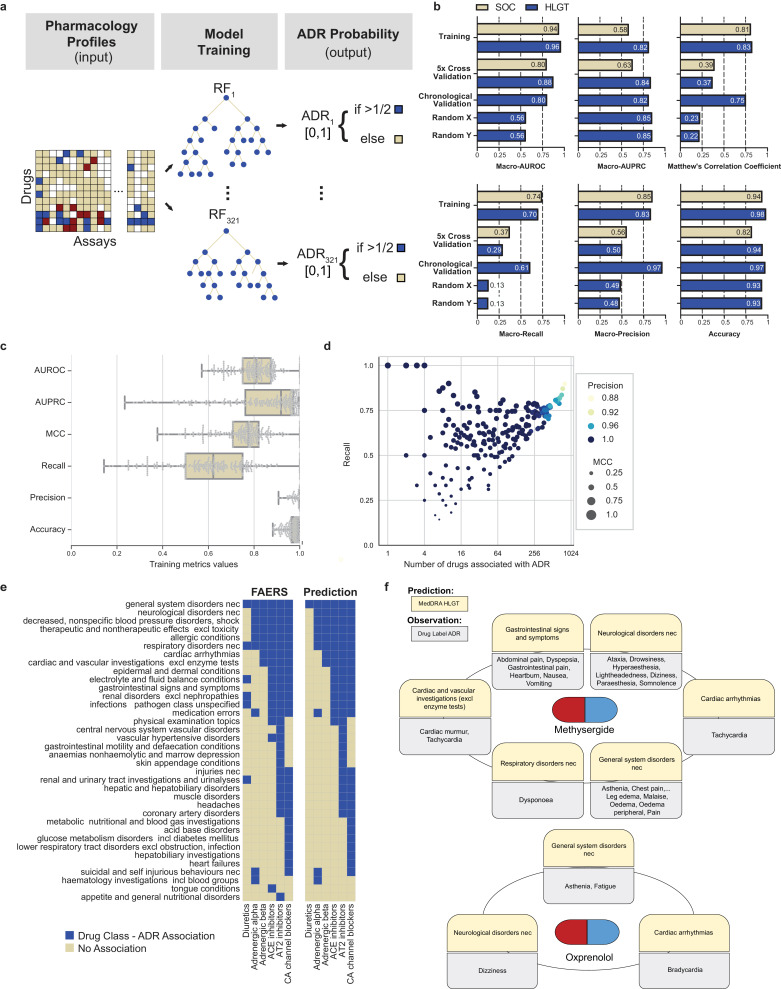
We deployed a machine learning approach to predict ADRs for a given drug from their in vitro secondary pharmacology profiles (Fig. 3a). We consider this a binary classification problem for each ADR independently because a given drug can cause multiple ADRs based on its possible engagement with multiple targets and because a single target may be associated with multiple ADRs. We discretized and one-hot encoded our “input” in vitro pharmacology assay data (AC50 values) into 3 classes: highly active (AC50 < 3 μM), active (3 μM ≤ AC50 ≤ 30 μM) and inactive (AC50 > 30 μM), which are in accordance with the dynamic range of all our assays (30 μM, except for BSEP as described below) and reflect commonly used ranges in the field [4]. Out of the 184 assays x 3 classes = 552 binary features to represent our assay information input data, those consisting of only 0 values were removed, resulting in 413 input features. These were used to predict 321 High Level Group Term (HLGT) ADRs or 26 System Organ Class (SOC) ADRs for each drug. The observed drug-ADR associations from FAERS, as described above, constitute the dependent variable that the model is learning. We constructed a unifying binary relevance random forest model, which consists of 321 random forest HLGT ADR models. The models were first trained and tested, using 5-fold cross validation where each fold is selected sequentially (Fig. 3b). We used 1329 drugs for model construction because these drugs had at least 1 adverse event report in FAERS Q4_2018. The remaining 805 drugs, which did not have any ADR reports, were excluded from training and cross validation. We confirmed that the distribution of drug classes in our training set (1329 drugs) is comparable to the distribution of drug classes in each 5-fold cross validation split (1063 drugs for training and 226 drugs for testing; Supplementary Fig. 1a, χ2-test: p-value > 0.99). Likewise, the observed drug-ADR associations forming the binary dependent variable of the random forest were also stratified between training and cross validation splits (Supplementary Fig. 1b). The model predictions are in probability format, which is used later for target-ADR predictions, and in boolean format (Fig. 3a), to enable assessment of model performance via the area under the receiver operating characteristic curve (macro-AUROC); the area under the precision-recall curve (macro-AUPRC); accuracy; macro-precision; macro-recall and Matthew's correlation coefficient (MCC), a performance measure that takes class imbalance into account (Fig. 3b). The unifying random forest model performance of SOC ADRs and HLGT ADRs using the full training set (1329 drugs) and the 5-fold cross validation sets (266 drugs, averaged) are depicted in Fig. 3b. Accuracy ranges from 0.82 to 0.98, macro-precision ranges from 0.5 to 0.85, macro-recall ranges from 0.29 to 0.74, MCC ranges from 0.37 to 0.83, and macro-AUROC ranges from 0.80 to 0.96. These model performances are higher than negative control random forest models trained and cross validated on randomized input (X) or output (Y) data, further confirming the predictive power of our models (Fig. 3b, Supplementary Table 4). Compared to SOC level (21 ADR terms), the finer grain HLGT level (321 ADR terms) had proportionally fewer drug-ADR associations; additionally, the performance of the HLGT and SOC models are comparable. We therefore proceeded with the HLGT level models for further investigation.
Fig. 3.
Application of the random forest model to characterize drug-ADR associations
a. Schematic representation of the machine learning approach. Using input data, which is a discretized AC50in vitro pharmacological profile, we built a separate random forest model for each adverse drug reaction (ADR) that predicts the probability of a drug causing that ADR. For training we used all drugs for which we could retrieve FAERS Q4_2018 adverse event reports (Ndrugs = 1329).
b. Summary statistics of overall model performance. We developed two unified random forest models based on two hierarchical levels of organ class specifications. The high level group term (HLGT; blue) unified random forest model consists of 321 ADR random forest models whereas the system organ class (SOC; yellow) unified random forest model consists of 26 ADR random forest models. The performance of the HLGT and SOC models is similar, except in a few cases when the HLGT model outperforms the SOC model. (MCC: Matthew's correlation coefficient, AUROC: area under receiver operating characteristic, AUPRC: area under precision recall curve). Training reflects performance after model training on all 1329 drugs (see A). 5-fold cross validation results are averaged over each fold (all metrics for each fold are detailed in Supplementary Table 4). Chronological validation reflects the performance of the HLGT level random forest models trained on FAERS reports up to Q4_2014 and tested on the observations up to Q2_2019. Random X and Y indicate the 5x cross validation performance of negative control HLGT level random forest models trained on randomly permuted input and output data, respectively.
c. Box plots indicating the distributions of the training performance metrics (as in B) for all random forest models of each individual HLGT ADR (NADRs = 266; center line, median; boxlimits, 1st and 3rd quartiles; whiskers, minimal and maximal value; points represent all data).
d. Scatter plot of the random forest models’ recall (all metrics as in C) as a function of number of associated ADRs, which served as positive training examples. Colours indicate model precision and circle size reflects the MCC.
e. ADR predictions for anti-hypertensive drugs with different pharmacological targets. For a set of 22 antihypertensive drugs, we visualized the association between the drugs and HLGT-level ADRs (left). Using the ADR random forest models, we predicted the differences in ADR associations between antihypertensive drugs representing various pharmacological targets (right; overall 36 of the HLGT terms are visualized). True negative predictions (285 HLGT-level ADRs) were omitted from this visualization.
f. Examples of model validation using methysergide and oxprenolol. The random forest model predicted associations of methysergide with 6 of 321 HLGTs (yellow) which were validated by comparison of ADRs from its drug label (grey) using the SIDER database. One or more of the ADRs corresponding to each HLGT category were confirmed in the drug label. .
For 55 of the 321 HLGT ADRs, the corresponding random forest models simply predicted zero for all drugs as mostly none (and at most 4) of the 1329 drugs with adverse event reports were associated with those ADRs (Supplementary Table 5, all ADRs with precision = NA). Intuitively, if too few drugs are reported to cause an ADR in FAERS, insufficient training data is available for our random forest to learn whether in vitro pharmacology drug profile could predict that particular ADR. Since these 55 models were not predictive, we did not consider them for further analyses. For the remaining 266 ADRs, we could determine performance metrics (Fig. 3c). Accuracy and precision were high, ranging between 0.9 and 1, whilst the recall and MCC range more widely (Fig. 3c). This variability occurs for ADRs that have only a few drugs associated with them (Fig. 3d). This indicates further that sparse positive training data, which causes a large class imbalance, generally results in reduced predictive power of the random forest model. As the number of associated drugs increases, the models learn to better distinguish true positives from false negatives, subsequently leading to an increase in recall and MCC values (Fig. 3d).
3.4. Predictive power of the random forest model for multiple FAERS reporting time periods
To test if our random forest model framework is sensitively dependent on the FAERS reporting period, we constructed new random forest models and performed 5-fold cross validations for both SOC and HLGT levels using FAERS data from 2 different time points: Q4_2014 (including all reports from January 2004 to December 2014) and Q2_2019 (including all reports from January 2004 to June 2019). For proper comparison, the model constructions and cross validations were identical to our above described “main” model based on FAERS Q4_2018. Overall, the performance metrics (accuracy, MCC, macro-precision, macro-recall, macro-AUROC) of both SOC and HLGT level models are comparable between Q4_2014, Q4_2018 and Q2_2019 (Supplementary Table 4). This analysis demonstrates that our random forest modeling framework has a comparable predictive power despite changes in the FAERS reporting time period; therefore, it is not sensitive to different versions of FAERS.
3.5. Chronological validation of predicted drug-ADR associations
To validate the predictive power of our random forest modeling framework further, we performed a chronological validation analysis, through identification of initial false predictions (false positives and false negatives) from the random forest model trained on FAERS Q4_2014 which was then validated using a dataset from the subsequent time period, 2015–2019. The random forest model trained on Q4_2014 data has 421 (0.1% of a total of N = 433,671 model predictions) false positive drug - ADR associations, i.e. based on a drug's pharmacology profile, the model predicted a probability > 0.5 (Fig. 3a) for an ADR even though there was no association observed from the adverse event reports up until 2014. However, when compared to the observed Q2_2019 FAERS data, which also include adverse event reports from the time period 2014–2019, 3.1% (13) of the false positives turned into observed drug-ADR associations (true positive), which is 4.4-fold more than expected by chance (χ2-test: p-value = 2×10–5). Similarly, the Q4_2014 random forest model made 8519 false negative predictions, of which 2.2% (184), 40-fold more than expected by chance (χ2-test: p-value < 10–16), turned into true negative predictions when compared to the Q2_2019 observed drug-ADR associations. Furthermore, the overall model performance metrics had also improved over time as compared to 5x cross validation on Q4_2014 (Supplementary Table 4) and Q4_2018 observations (Fig. 3b). This analysis indicates that significant proportions of our model predictions on drug-ADR associations that were initially “false predictions” became “true predictions” through accumulation of new adverse events reports over time.
3.6. Random forest model predicts expected ADR profiles for anti-hypertensive drugs
As another demonstration of model validation, we analysed the ADR profiles of 6 subclasses of antihypertensive drugs: adrenergic alpha, adrenergic beta, ACE inhibitors, angiotensin AT2 inhibitors, calcium channel blockers and diuretics (Supplementary Table 6). The signature of the anti-hypertensive drug subclass represents a set of ADRs that were common to all drugs in this subclass. Each antihypertensive drug subclass has a unique ADR fingerprint in the Q4_2018 FAERS version which was closely predicted by our random forest model (Fig. 3e). The accuracy ranged from 0.984 to 1, with perfect specificity and precision (Supplementary Table 7). The sensitivity ranged from 0.882 to 1, except for the diuretics sub-class, which had a sensitivity of 0.167. This may be because diuretics target the kidney, and not the cardiovascular system as the rest of the anti-hypertensive drugs do. Of note, the adrenergic alpha and adrenergic beta receptor subclasses maintain distinct profiles in the predicted data. Specifically, the model correctly predicts that adrenergic alpha receptor drugs are associated with suicidal and self injurious behaviors, which has been reported in the literature [39,40].
3.7. Random forest model validation through comparison with drug label ADRs
To demonstrate the predictive power of our random forest model on a test set of drugs that were not used for model construction, we utilized the model to predict drug-ADR associations for 805 drugs that did not have any reported ADRs in the FAERS Q4_2018 version, either because there was no match with the drug name or there were no ADR reports for that drug submitted to FAERS by October 2018. For validation, we queried the Side Effect Resource (SIDER) database [24], which contains drug-ADR pairs extracted from FDA drug labels by text mining [24]. Drug labels are generally informed by FAERS and other sources. For these 805 drugs without evidence from FAERS, we obtained 95 drug matches in SIDER, which were further reduced to 75 drugs that did not share active ingredients with drugs in the training set. Overall, 57% of positive drug-ADR pairs (i.e. drugs where the model predicts ADRs) were reported in SIDER, compared to 9% of negative pairs (N = 24,075; χ2-test: p-value < 10–16; Supplementary Table 8). For instance, methysergide, a 5-HT receptor antagonist used to treat migraine and cluster headaches, has predicted ADRs from 6 HLGT categories, all of which are supported by specific ADRs from SIDER (Fig. 3f). “Cardiovascular disorders with murmurs” appears in the Warnings and Precautions section of the label. Other adverse events under gastrointestinal symptoms and CNS symptoms from SIDER were confirmed in the Adverse Events section. Oxprenolol, a lipophilic beta blocker used for treating angina pectoris, abnormal heart rhythms and high blood pressure, has predicted ADRs from 3 HLGT categories. The specific SIDER ADRs of bradycardia, dizziness and asthenia were also confirmed in the label from the Electronic Medicines Compendium (https://www.medicines.org.uk/emc/product/3235; accessed 09/11/2019). Overall, our random forest model proves to be a powerful tool to predict both on- and off-target related drug-ADR associations from in vitro pharmacological drug profiles.
3.8. Random forest performance and validation on OMOP benchmark
To assess the predictive power of our random forest model further, we compared our drug-ADR associations against the OMOP benchmark [26], which consists of 399 drug-ADR pairs with a binary ground truth (association or no association) as determined through evaluation of different information sources (including FAERS) by domain experts [26]. We mapped the 4 ADRs represented in OMOP (acute myocardial infarction, renal failure acute, liver disorder and gastrointestinal haemorrhage) to their respective HLGT parent classes. For evaluation, we included all drug-ADR pairs that were present in OMOP and also had available model predictions (Supplementary Fig. 2, Supplementary Table 9). For comparison, we also applied this benchmark procedure to drug-ADR association predictions from ERAM [8,[36], [37], [38]) and our above described binomial model, which both rely on the FAERS adverse event reports to determine the statistical significance of drug-ADR pairs (see Methods for details). As described above, the binomial model drug-ADR associations were used as output training data for our random forest model. Consistently, our trained random forest evaluated on the RF training set drugs performed largely on par with our binomial model and ERAM [8,[36], [37], [38]]. Interestingly, for the smaller test set of drugs without FAERS reports, but nevertheless present in the OMOP benchmark, our RF model predicted the ground truth for 22 drug-ADR pairs with a high AUROC and AUPRC of 0.98 and 0.80, respectively (Supplementary Fig. 2). These results indicate that our trained RF model can generalize to predict ADR associations based on only a drug's in vitro profile. This is of particular interest to efforts toward the safety assessment of preclinical drug candidates during the drug discovery process.
3.9. Random forest model predicts 221 target-ADR associations
To predict which target genes are associated with which ADRs, we utilized the Gini importance score to rank features for their importance in random forest models for each ADR (Fig. 4a). For a given ADR, we selected assays that had multiple AC50 features represented in the top 5% of Gini scores ranking (see Methods for detail). We then generated ADR probability predictions for an in silico compound that is assumed to target only the selected assay with an AC50 value corresponding to a represented feature. We also assumed no available data for all other assays. Using this in silico AC50 profile as an input to the ADR model, we could generate the ADR probability. By assessing differences in ADR probabilities (two sample t-test, FDR corrected p-value < 0.1) between different AC50 classes, e.g. highly active (0–3 μM) vs inactive (>30 μM), we predict positive or negative correlations, collectively termed associations, between the selected target assay and ADR. Unsurprisingly, some ADRs did not generate any target associations.
Fig. 4.
Random forest model predicts target-ADR associations.
a. Schematic outline of the in silico ADR-target predictions. For an ADR of interest, we determined the top 5% of features from the corresponding trained random forest model, ranked according to their Gini importance scores, which measures their contribution to the predictive power of the model. If at least two features (e.g. as depicted: highly active and inactive) from the same target assay are within that top 5%, we determined the ADR probabilities for the simulated cases where an in silico compound would target those assay AC50 classes only. The ADR probabilities of those simulated cases can then be compared to determine the concentration dependence of the ADR probability. If there is a non-zero correlation between AC50 values and ADR probabilities, we conclude that there is an association between the respective ADR and target. For full details, see the Methods.
b. Heatmap showing the resulting 221 predicted target-ADR associations (blue). Target (gene symbol) assays are listed alphabetically (horizontal), and HLGT ADRs (vertical) are grouped according to their parent SOC level (as detailed in Fig. 2c). For a full description of all target-ADR associations and their ADR probabilities, see Supplementary Table 10.
c. Scatter plot of each target (assay, N = 184) showing the number of ADR associations as a function of number of assayed drugs.
To find biologically meaningful associations, we first filtered out HLGT terms belonging to SOC classes that are not specific to human body parts or only procedural or intervention related (see Methods for detail). Secondly, we filtered out terms that fall under the SOC class neoplasms, since genes are often severely misregulated in cancers and therefore not representative of neoplasm-related ADRs in the organ where the tumour resides. After filtering, we found 221 statistically significant target-ADR associations (Fig. 4b, full details including p-values in Supplementary Table 10); 51 out of 184 target assays and 132 out of 321 ADRs are represented (Fig. 4b). The assay class distribution of these 51 targets, represented among the 221 predicted target-ADR associations, is similar to the class distribution of all target assays (Supplementary Fig. 1b, χ2-test: p-value = 0.09). This demonstrates that our algorithm does not bias towards certain target classes. In the following sections, we investigate these 221 target-ADR associations in more detail.
3.10. Systematic literature validation of target-ADR associations
To validate our ADR-target predictions, we performed a systematic literature co-occurrence analysis. First, we mapped all genes corresponding to the assays and HLGT level ADRs to their respective MeSH terms (Supplementary Table 11). Next, we queried PubMed for the publication identifiers linked to these MeSH terms and determined the number of publications that corresponded to both a gene and HLGT term (i.e. co-occurrence). We found at least one co-occurrence publication for 66% (145) of 219 predicted unique gene-HLGT MeSH pairs, which was higher (Fisher Exact test: odds ratio=1.8, p-value=6×10–5) than for all possible negative unique gene-HLGT pairs (N = 26,705). In order to control for the fact that some ADRs and genes are studied more intensively than others, we also compared our set of positive predictions to a negative control set (N = 4890) formed by permuted pairs from the positive set and obtained similar results (Fisher Exact test: odds ratio=1.5, p-value=3×10–3). Furthermore, as quantified by the co-occurrence “lift” over the reporting probability when assuming independence, (see Methods for details), we found 4-fold higher co-occurrence median lift values for our predictions compared to all negative pairs (Mann Whitney U test: p-value=2×10–5), and 3-fold higher lift than permuted negative pairs (Mann Whitney U test: p-value=3×10–4). We conclude that our target-ADR identification method provides association predictions that are supported by the literature in higher proportion than random selection of target-ADR pairs.
3.11. Evidence for targets that are predicted to cause cardiovascular-related ADRs
To further validate our model's ability to predict target-ADR associations, we investigated a group of cardiovascular ADRs. We found that hERG binding was associated with cardiac arrhythmias and heart failure (Table 1). hERG encodes for a subunit of the cardiac potassium ion channel and contributes to cardiac electrical activity, which is necessary to regulate the heartbeat. The mechanism of action for drug-induced arrhythmias by blocking hERG has been described in numerous human [41] and animal studies [42], as well as structural modeling [43] studies (Table 1). Consistently, our systematic PubMed queries found 753 co-occurrence publications in support of this predicted association and 6 co-occurrences for hERG binding increasing the risk of heart failure. We did not find an ADR probability associated within the range of 0–3 μM AC50 of hERG binding, likely because such strong binding to hERG is a common reason for deprioritizing drug candidates in development [44].
Table 1.
Predicted associations between targets and cardiac ADRs. High Level Group Terms (HLGT; MedDRA) associations with targets and Adverse Drug Reaction (ADR) probability in three concentration ranges (third column). Evidence of the ADR-target pairs were obtained from peer reviewed publications (fourth column). The number of publications linked to both an HLGT ADR and target gene was obtained via a systematic literature co-occurrence analysis (fifth column). hERG: human Ether-a-go-go-Related Gene associated potassium channel; PDE3: phosphodiesterase-3 enzyme; GR: glucocorticoid receptor; AdT: Adenosine transporter; COX-2: cyclooxygenase enzyme, type 2.
| Cardiac Disorder HLGT | Target | ADR Probability |
Literature evidence | Co–occurrence (number) | ||
| 0–3 μM | 3–30 μM | >30 μM | human (h), animal (a), in vitro (v) | |||
| cardiac arrhythmias | hERG (Binding) | – | 0.03 | 0.002 | h [41] a [42] v [43] | 753 |
| cardiac valve disorders | PDE3 | 0.05 | – | 0 | h [45,46] | 3 |
| heart failures | hERG (Binding) | – | 0.005 | 0 | h [75] | 6 |
| myocardial disorders | GR (Binding) | 0.02 | – | 0.005 | h [48,49] | 8 |
| pericardial disorders | AdT | – | 0.01 | 0 | a [47] | 0 |
The model predictions also suggest that PDE3 inhibition is associated with cardiac valve disorders (Table 1, 3 co-occurrence publications). PDE3 inhibition is used clinically to treat dilated cardiomyopathy [45], which encapsulates valvular heart disorder. However, the PDE3 therapeutic window is narrow, partially due to complex signaling networks [46], and careful dosing is required to avoid increased mortality in response to treatment.
Table 3.
Predicted ADR associations with inhibition of the Bile Salt Export Pump (BSEP) transporter (detailed legend in Table 1).
| HLGT | Target | ADR Probability |
Literature evidence | Co–occurrence (number) | ||
| 0–3 μM | 3–30 μM | >30 μM | human (h); animal (a) | |||
| central nervous system vascular disorders | BSEP | – | 0.09 | 0.008 | (for BSEP and bile acid) a [79] | 2 |
| foetal complications | BSEP | 0.01 | – | 0 | h [62] | 7 |
| pregnancy labor delivery and postpartum conditions | BSEP | – | 0.1 | 0 | h [63] | 0 |
| lipid metabolism disorders | BSEP | – | 0.2 | 0 | h [80,81] | 5 |
| thyroid gland disorders | BSEP | – | 0.07 | 0 | a [82,83] | 1 |
| upper respiratory tract disorders excl infections | BSEP | 0.1 | – | 0 | h [84] a [85] | 0 |
| urolithiases | BSEP | – | 0.07 | 0 | h [86] | 0 |
| 0–30 μM | 30–300 μM | >300 μM | ||||
| hepatic and hepatobiliary disorders | BSEP | – | 0.2 | 0.09 | h [60] | 354 |
Furthermore, our model predicts that adenosine transporter (AdT) inhibition increases the risk of pericardial disorders (Table 1). For this scenario, we did not find direct supporting evidence in the literature, however there is evidence that disturbed adenosine homoeostasis in pathological cardiac conditions could result in pericardial effusion or pericarditis [47].
The model suggests that glucocorticoid receptor (GR) binding is more likely to lead to myocardial disorders if the drug has high affinity for GR (Table 1, 8 co-occurrence publications). This is supported by the finding that glucocorticoid treatment of patients with rheumatoid arthritis increased the risk of myocardial infarction [48]. Furthermore, it is known that dysregulation of glucocorticoids can give rise to cardiotoxicity [49].
Taken together, this investigation of genes associated with cardiovascular ADRs confirms the well-known association of hERG with cardiac arrhythmia, and also highlights ADR associations that would merit further experimental investigation.
3.12. COX-2, PDE3, and hERG associations with kidney related ADRs
Another important class of ADRs involve the kidney (Fig. 4b, label: renal). We found COX-2 associated with nephropathies (Table 2), which has been well recognized (398 co-occurrence publications) and evidenced previously [50], [51], [52]. Interestingly, another model prediction is PDE3 sensitivity correlating with congenital renal and urinary tract disorders (Table 2). According to a mouse model study [53], PDE3 inhibition could be a contributing factor in Polycystic Kidney Disease (PKD), as PDE3 protein levels are downregulated in PKD compared to healthy control kidneys. Lastly, we found an unexpected association between hERG and renal disorders (excluding nephropathy) (Table 2). One study has found a loss of hERG function in renal cell carcinoma [54]. In humans, hERG expression in the kidney is much lower than in the heart [55]. Therefore, we conclude that a link between hERG and renal disorders remains a prediction that warrants further investigation.
Table 2.
Predicted renal ADR - target associations (detailed legend in Table 1).
| Renal Disorder HLGT | Target | ADR Probability |
Literature evidence | Co–occurrence (number) | ||
| 0–3 μM | 3–30 μM | >30 μM | human (h); animal (a), in vitro (v) | |||
| nephropathies | COX-2 | 0.003 | – | 0 | h [50] a [51,52] | 398 |
| renal and urinary tract disorders congenital | PDE3 | 0.004 | – | 0 | h [66,76] a [53,77] | 0 |
| renal disorders excl nephropathies | hERG (Binding) | – | 0.01 | 0.0007 | h [54] a [78] | 2 |
3.13. PDE3 and nuclear hormone receptors AR, ERa, and PR are overrepresented in ADR associations
To investigate if the number of different drugs tested for a target assay is predictive to the number of ADRs associated with that target (Fig. 4c), we calculated their Spearman correlation coefficient and found a moderate correlation (ρ=0.5; Fig. 4c). However, some targets had considerably more associated ADRs than other targets that were tested a similar number of times, indicating that more frequently performed assays do not necessarily result in a higher number of associated ADRs (Fig. 4c). Out of all target assays, PDE3 was associated with the most ADRs (40, Fig. 4c), falling in a wide range of SOC classes (Fig. 4b, Supplementary Table 10). Furthermore, nuclear hormone receptors for androgen (AR), oestrogen (ERa) and progesterone (PR) binding assays also have disproportionately many ADR associations, compared to their frequency of testing (Fig. 4c). As expected, AR (7/14 ADRs), ERa (9/10 ADRs) but not PR (0/17 ADRs) are associated with sexual reproductive organ- and pregnancy-related ADRs (Fig. 4b, Supplementary Table 10). Androgen is produced in the adrenal gland [56] and we predict a link between AR with adrenal gland disorders, with evidence in mouse studies [57]. Interestingly, the model predicted 6 ocular ADRs associated to PR, including vision disorders, anterior eye structural change (deposit and degeneration), infections, irritations and inflammations and structural changes (Fig. 4b, Supplementary Table 10), for which we could find supporting evidence [58].
3.14. GABAA receptor associations with psychoactive ADRs
GABAA receptor is the primary target of benzodiazepines (BZD), a drug class known to be psychoactive with potential of addiction [59]. Consistently, our model predicts that this ligand-gated chloride ion channel assay is associated with 14 ADRs, 13 of which are neurologically and psychiatrically related, including disturbances in thinking and perception, sleep disorders, depression and suicidal behaviors (Fig. 4b, Supplementary Table 9).
3.15. Bile salt export pump BSEP associations with ADRs in various organs
BSEP, encoded by ABCB11 and a member of the superfamily of ATP-binding cassette transporters, is most highly expressed in the liver [55]. Drugs that target BSEP are often associated with hepatotoxicity [60]. However, initially, we did not find a BSEP association with hepatic and hepatobiliary disorders. To investigate this false negative prediction, we note that the dynamic range of all our assays extend to 30 uM, except for the BSEP assay, which specifically extends up to 300 μM because the first pass effect for orally delivered drugs results in high concentrations in the liver [61]. As a result, with our default class boundaries, most of our BSEP data falls into the ‘inactive’ (>30 uM class). Consistently, the BSEP inactive feature had the highest Gini score for this HLGT term, while its two active features had much lower Gini scores, falling outside of the top 5%. To take the extended dynamic range into account, we altered the BSEP assay class boundaries to 0–30 μM, 30–300 μM and >300 μM and retrained the random forest model. In this case, we did find BSEP associated with hepatic and hepatobiliary disorders (Table 3, 354 publication co-occurrences), according to our association criteria (Fig. 4a). We repeated this procedure whilst replacing the first class boundary (30 μM) with 100 μM and found the same association again, indicating the robustness of our results. Interestingly, with our original AC50 discretisations (Fig. 1d), we found BSEP associated with 7 other ADRs from various organ classes (Table 3), much more than other targets that were assayed at a similar frequency (Fig. 4c). This suggests that compounds potent against BSEP (AC50 < 30 μM) could cause adverse effects in addition to hepatotoxicity, which already occurs at lower potency. We found BSEP associated with urolithiasis and with disorders of the thyroid gland, upper respiratory tract disorders (excl infections), lipid metabolism and central nervous system (Table 3). Since BSEP expression is much lower in these organs [55], we searched the literature for evidence including a substrate of BSEP, bile acid. We could find previous studies linking bile acid to these disorders (Table 3), which suggests an indirect relation between BSEP and these ADRs through bile acid metabolism. Lastly, we found BSEP associated with foetal complications and pregnancy conditions (Table 3), both supported through prior studies that link BSEP with transient neonatal cholestasis and intrahepatic cholestasis of pregnancy, respectively [62,63].
4. Discussion
In this study we have taken a machine learning approach to predict human ADRs from the in vitro secondary pharmacology profiles of a large number of marketed and withdrawn drugs. Several prior studies focus on predicting ADRs directly from chemical drug structure [64,65]. However, under the common assumption that our in vitro pharmacology adequately reflects the in vivo activity of compounds, utilizing this functional information on targeting of common (off) targets represents a viable alternative to bridge the complex relationship between drugs and their effects in the human body [4].
Our random forest model performance metrics are good considering the sparse coverage (2134 drugs) over a large input space (3184 possibilities) and partial overlap with ADR reporting for these drugs, making ADR occurrence prediction effectively a one shot learning task. Importantly, our model performances were strong enough to discover drug-ADR and biologically meaningful target-ADR associations. To determine the target-ADR associations, we utilized all available input data for model training and made use of Gini scores to robustly select relevant features for ADR probability predictions. We rigorously validated our model predictions with multiple independent analyses (e.g. chronological validation on drug-ADR associations and systematic literature validation on target-ADR associations). Our novel method for target-ADR associations was able to recapitulate well recognized causal relations, such as hERG with cardiac arrhythmias. For others, we were able to find literature evidence in animal or in vitro studies but our study is, to our knowledge, a first in human report. Another fraction of target-ADR associations represents predictions of novel, unexpected or little known associations, such as Adenosine Transporter (AdT) and pericardial disorders, for which we could find little evidence other than our analysis of adverse event reports. Similar to genome-wide association studies (GWAS), our quantitative methodology extracts statistically significant relations from human population data. With this framework in mind, our 221 associations form a rich resource that can be used for further mechanistic studies in the drug discovery process.
Our random forest model is agnostic to molecular mechanisms; therefore, resulting associations could arise from indirect regulation. A likely example is the bile transporter BSEP, which is associated with numerous ADRs, although it is most highly expressed in the liver and kidney. We have related our findings to evidence that misregulation of its substrate, bile acid, could result in disorders related to kidney stones, lipid metabolism, thyroid gland, respiratory system, and central nervous system. This also indicates the strength of our approach, which can relate genes to physiological processes unbiasedly in humans, without any interventions or costly large scale population studies, but solely with voluntary adverse event reporting.
Some of the predicted target-ADR associations could be hard to validate, such as the PDE3 enzyme association with congenital renal disorders association. While the association is valid, the modality has to be clarified: PDE3 inhibitors are proposed to ameliorate certain forms of chronic kidney disease [66], instead of causing it. Thus, predictions of congenital disorders should be considered but confirmed by checking the modality of the effects.
While we recommend this approach to find target-ADR associations to impact safety awareness in drug discovery, we are also aware of the limitations. Firstly, targets in the in vitro pharmacology panel cover a fraction of the biological target space and not all drugs were tested in all assays. We recognize that 47% of all targets belong to the GPCR target family with limited representation of other therapeutic or ADR-associated targets such as ion channels and kinases. However, our model predictions are not biased towards the GPCR target family; the target classes of 51 targets in 221 target-ADR association predictions have a similar distribution compared to all target classes in our input data (Supplementary Fig. 3). Also, data are influenced by prior knowledge; for example, more than 87% of all drugs in the set were tested for hERG activity. High affinity (lower AC50 value) for hERG is associated with higher probability for QT prolongation for human and non-human preclinical species [41,42]. As discussed earlier, there are not many drugs with a hERG AC50 value in the highly active class (0–3 μM), which is a commonly encountered roadblock for drug candidates to progress towards clinical trials [44]. Only about 10% of all drugs fall into the highly active class in our assay data. To limit feature engineering, our AC50 discretization into three classes (Fig. 1d) was kept uniform across all assays. Notably for the BSEP assay only, the dynamic range extends up to 300 μM and as a result most of our data falls into the ‘inactive’ (>30 μM) class. Consequently, we initially did not find the expected association with hepatotoxicity. We rectified this by reclassifying the BSEP assay data according to levels required for hepatotoxicity of BSEP inhibition [67,68] and indeed recovered the expected association.
Secondly, in vitro potency is an initial marker of clinical effect, and does not take into account prolonged dosing, comorbidity or pharmacokinetic/pharmacodynamic (PK/PD) relationships (e.g. therapeutic window). For 9 of 184 assays, non-human proteins were assayed (e.g. rat brain was used as a source for the benzodiazepine receptor) which may not be a direct correlate of the human protein. One way of further improvement of our approach is to include additional occupancy parameters and PK/PD components for higher precision and enhanced predictive value.
Lastly, in the FAERS database, drug-ADR associations may be mislabeled, e.g. anti-hypertensives are often reported as associated with hypertension as an ADR, rather than as the indication. Additionally, the FAERS database does not contain information on the total number of patients exposed to a particular drug, nor is it necessarily a reflection of the true incidence or frequency of ADRs. These and other limitations are discussed by Maciejewski et al. [35] with suggestions and methodology for further refinement of the FAERS database curation and maintenance. We modeled ADRs at the MedDRA HLGT level instead of the finer grain Preferred Term level, because this reduces the sparsity of drug-ADR associations and enables the random forest model to learn. The HLGT level ADRs provide sufficient physiological detail to advance human biology and drug discovery, in particular during the lead optimization phase of drug discovery, where off-target mitigation is possible and should be a priority. This can contribute to the generation of safer clinical candidates and accelerate the drug development process, also acknowledged by the FDA.
We investigated one-to-one associations between targets and ADRs because these relationships are biologically meaningful and have a high utility in preclinical drug development. Given this objective, we considered each ADR to be independent from one another. However, in some cases, a given ADR can be a prerequisite for others (e.g. hypotension leading to reflex tachycardia), as considered previously in the context of drug-drug interaction predictions [10]. We leave a model extension to incorporate ADR dependencies as future work. For target-ADR associations, we utilized our random forest model for a single drug at a time. Our model can be repurposed to predict possible ADRs from combination drug therapies and likelihood of drug-drug interactions. In principle, this can be extended for combination therapies by merging the in vitro data from the individual compounds and the predictions can be validated by querying Offside and Twosides databases [9]. Similarly, our model can be utilized for drug repositioning and repurposing, using similar drug-ADR and target-ADR profiles. To a certain extent, our gene target-ADR associations resemble GWAS results, but for adverse reactions manifesting in patients (ADRs) instead of quantitative traits. These associations could help (de)prioritize drug candidates in the preclinical development stage based on their predicted side effects. This preclinical computational assessment may enable a cost effective approach to reduce the high clinical drug failure rate, which is predominantly caused by safety issues and poses a large financial burden. Secondly, our target-ADR associations could help advance human biology, as they predict the human in vivo effects of perturbing a protein target. Such experiments are often infeasible in humans due to ethical reasons, whilst equivalent experiments in animal models do not always mirror the human response. Our approach provides an alternative source of information for the targets we have investigated. In conclusion, our random forest model and the target-ADR associations provide a validated, comprehensive resource to support drug discovery and development as well as future human biology studies.
Declaration of Competing Interest
None.
Acknowledgments
Data availability
Data is made available in Supplementary Tables 1–11 and on GitHub (https://github.com/samanfrm/ADRtarget).
Code availability
Code to query FAERS and PubMed, to construct the random forest models and identify the target-ADR associations is made available on GitHub (https://github.com/samanfrm/ADRtarget).
Acknowledgments
We are grateful to Mirjam Trame and Andy Stein for giving us the opportunity to participate in the 2018 Novartis Quantitative Sciences Academia-to-Industry Hackathon organized at the Novartis Institutes for Biomedical Research. We also thank Changchang Liu and Xinrui (Sandy) Zou for their contributions to the project at the Hackathon.
Role of the funding source
This study was not supported by any formal funding bodies.
Author contributions
R.I., S.A., A.X.C., S.F., B.K., D.A. and L.U. conceived the study. D.A., A.F. and L.U. provided the Novartis in vitro pharmacology data, advice and mentorship. S.A., R.I., A.X.C., S.F., B.K., W.D.M. and J.S. performed data analysis. S.A. developed the random forest modeling. R.I. developed the formalism for target-ADR association inference. S.F., R.I. and A.X.C. developed the query of OpenFDA. J.S. performed the SIDER analysis. S.F., J.S. and R.I. performed the systematic PubMed query. R.I., S.A. and A.X.C. wrote the paper and designed the figures with input from all the authors.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.102837.
Contributor Information
Robert Ietswaart, Email: robert_ietswaart@hms.harvard.edu.
Seda Arat, Email: seda8arat@gmail.com.
Jeffrey J. Sutherland, Email: jeffrey.sutherland@novartis.com.
Laszlo Urban, Email: laszlo.urban@novartis.com.
Appendix. Supplementary materials
References
- 1.Institute of medicine, committee on quality of health care in America . National Academies Press; Washington (DC): 2000. To err is human: building a safer health system. [PubMed] [Google Scholar]
- 2.Lazarou J., Pomeranz B.H., Corey P.N. Incidence of adverse drug reactions in hospitalized patients: a meta-analysis of prospective studies. JAMA. 1998;279(15):1200–1205. doi: 10.1001/jama.279.15.1200. [DOI] [PubMed] [Google Scholar]
- 3.Weiss A.J., Freeman W.J., Heslin K.C., Barrett M.L. Adverse drug events in US hospitals, 2010 versus 2014. HCUP Stat Brief. 2018;234 https://www.hcup-us.ahrq.gov/reports/statbriefs/sb234-Adverse-Drug-Events.jsp [Google Scholar]
- 4.Lounkine E., Keiser M.J., Whitebread S., Mikhailov D., Hamon J., Jenkins J.L. Large-scale prediction and testing of drug activity on side-effect targets. Nature. 2012;486(7403):361–367. doi: 10.1038/nature11159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowes J., Brown A.J., Hamon J., Jarolimek W., Sridhar A., Waldron G. Reducing safety-related drug attrition: the use of in vitro pharmacological profiling. Nat Rev Drug Discov. 2012;11(12):909–922. doi: 10.1038/nrd3845. [DOI] [PubMed] [Google Scholar]
- 6.Witek R.P., Bonzo J.A. Perspective on in vitro liver toxicity models. Appl In Vitro Toxicol. 2018;4(3):229–231. [Google Scholar]
- 7.Whitebread S., Hamon J., Bojanic D., Urban L. Keynote review: in vitro safety pharmacology profiling: an essential tool for successful drug development. Drug Discov Today. 2005;10(21):1421–1433. doi: 10.1016/S1359-6446(05)03632-9. [DOI] [PubMed] [Google Scholar]
- 8.Dumouchel W. Bayesian data mining in large frequency tables, with an application to the FDA spontaneous reporting system. Am Stat. 1999;53(3):177–190. [Google Scholar]
- 9.Tatonetti N.P., Ye P.P., Daneshjou R., Altman R.B. Data-driven prediction of drug effects and interactions. Sci Transl Med. 2012;4(125):125ra31. doi: 10.1126/scitranslmed.3003377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zitnik M., Agrawal M., Leskovec J. Modeling polypharmacy side effects with graph convolutional networks. Bioinformatics. 2018;34(13):i457–i466. doi: 10.1093/bioinformatics/bty294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Portanova J., Murray N., Mower J., Subramanian D., Cohen T. aer2vec: distributed representations of adverse event reporting system data as a means to identify drug/side-effect associations. AMIA Annu Symp Proc. 2019;2019:717–726. [PMC free article] [PubMed] [Google Scholar]
- 12.Basile A.O., Yahi A., Tatonetti N.P. Artificial intelligence for drug toxicity and safety. Trends Pharmacol Sci. 2019;40(9):624–635. doi: 10.1016/j.tips.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ietswaart R., Gyori B.M., Bachman J.A., Sorger P.K., Churchman L.S. GeneWalk identifies relevant gene functions for a biological context using network representation learning. bioRxiv. 2019; Available from: https://www.biorxiv.org/content/10.1101/755579v2 [DOI] [PMC free article] [PubMed]
- 14.Noorbakhsh J., Farahmand S., Pour A.F., Namburi S., Caruana D., Rimm D., et al. Deep learning-based cross-classifications reveal conserved spatial behaviors within tumor histological images. bioRxiv. 2020. Available from: https://www.biorxiv.org/content/10.1101/715656v2 [DOI] [PMC free article] [PubMed]
- 15.Stokes J.M., Yang K., Swanson K., Jin W., Cubillos-Ruiz A., Donghia N.M. A deep learning approach to antibiotic discovery. Cell. 2020;180(4):688–702.e13. doi: 10.1016/j.cell.2020.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen P.A., Born D.A., Deaton A.M., Nioi P., Ward L.D. Phenotypes associated with genes encoding drug targets are predictive of clinical trial side effects. Nat Commun. 2019;10(1):1579. doi: 10.1038/s41467-019-09407-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu K., Sun X., Jia L., Ma J., Xing H., Wu J. Chemi-net: a molecular graph convolutional network for accurate drug property prediction. Int J Mol Sci. 2019;20:3389. doi: 10.3390/ijms20143389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ekins S. Predicting undesirable drug interactions with promiscuous proteins in silico. Drug Discov Today. 2004;9(6):276–285. doi: 10.1016/S1359-6446(03)03008-3. [DOI] [PubMed] [Google Scholar]
- 19.3rd Lynch JJ, TR Van Vleet, Mittelstadt S.W., Blomme E.A.G. Potential functional and pathological side effects related to off-target pharmacological activity. J Pharmacol Toxicol Methods. 2017;87:108–126. doi: 10.1016/j.vascn.2017.02.020. [DOI] [PubMed] [Google Scholar]
- 20.Hamon J., Whitebread S., Techer-Etienne V., Le Coq H., Azzaoui K., Urban L. In vitro safety pharmacology profiling: what else beyond hERG? Future Med Chem. 2009;1(4):645–665. doi: 10.4155/fmc.09.51. [DOI] [PubMed] [Google Scholar]
- 21.Mirams G.R., Cui Y., Sher A., Fink M., Cooper J., Heath B.M. Simulation of multiple ion channel block provides improved early prediction of compounds’ clinical torsadogenic risk. Cardiovasc Res. 2011;91(1):53–61. doi: 10.1093/cvr/cvr044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang Ll-H, He Q.-.S., Liu K., Cheng J., Zhong M.-.D., Chen Ll-S. ADReCS-target: target profiles for aiding drug safety research and application. Nucleic Acids Res. 2018;46(D1):D911–D917. doi: 10.1093/nar/gkx899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farahmand S., Riley T., Zarringhalam K. ModEx: a text mining system for extracting mode of regulation of transcription factor-gene regulatory interaction. J Biomed Inform. 2020;102 doi: 10.1016/j.jbi.2019.103353. [DOI] [PubMed] [Google Scholar]
- 24.Kuhn M., Letunic I., Jensen L.J., Bork P. The SIDER database of drugs and side effects. Nucleic Acids Res. 2016;44(D1):D1075–D1079. doi: 10.1093/nar/gkv1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.U.S. Food and Drug Administration (FDA). Questions and answers on FDA's adverse event reporting system (FAERS). Available from: https://www.fda.gov/drugs/surveillance/fda-adverse-event-reporting-system-faers
- 26.Stang P.E., Ryan P.B., Racoosin J.A., Overhage J.M., Hartzema A.G., Reich C. Advancing the science for active surveillance: rationale and design for the observational medical outcomes partnership. Ann Intern Med. 2010;153(9):600–606. doi: 10.7326/0003-4819-153-9-201011020-00010. [DOI] [PubMed] [Google Scholar]
- 27.Oliveira J.L., Lopes P., Nunes T., Campos D., Boyer S., Ahlberg E. The EU-ADR Web Platform: delivering advanced pharmacovigilance tools. Pharmacoepidemiol Drug Saf. 2013;22(5):459–467. doi: 10.1002/pds.3375. [DOI] [PubMed] [Google Scholar]
- 28.Chen A.W. Predicting adverse drug reaction outcomes with machine learning. Int J Commun Med Public Health. 2018;5(3):901–904. [Google Scholar]
- 29.Canham S.M., Wang Y., Cornett A., Auld D.S., Baeschlin D.K., Patoor M., et al. Systematic chemogenetic library assembly. bioRxiv. 2020. Available from: https://www.biorxiv.org/content/10.1101/2020.03.30.017244v1 [DOI] [PubMed]
- 30.Takarabe M., Kotera M., Nishimura Y., Goto S., Yamanishi Y. Drug target prediction using adverse event report systems: a pharmacogenomic approach. Bioinformatics. 2012;28(18):i611–i618. doi: 10.1093/bioinformatics/bts413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mozzicato P. MedDRA. Pharm Med. 2009;23(2):65–75. [Google Scholar]
- 32.Hauser A.S., Attwood M.M., Rask-Andersen M., Schiöth H.B., Gloriam D.E. Trends in GPCR drug discovery: new agents, targets and indications. Nat Rev Drug Discov. 2017;16(12):829–842. doi: 10.1038/nrd.2017.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.for Drug Statistics Methodology WCC. Guidelines for ATC classification and DDD assignment. Who Oslo; 2005.
- 34.U.S. Food and Drug Administration (FDA). openFDA. Available from: https://open.fda.gov
- 35.Maciejewski M., Lounkine E., Whitebread S., Farmer P., DuMouchel W., Shoichet B.K. Reverse translation of adverse event reports paves the way for de-risking preclinical off-targets. Elife. 2017;6 doi: 10.7554/eLife.25818. e25818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fram D.M., Almenoff J.S., DuMouchel W. Proceedings of the ninth ACM SIGKDD international conference on knowledge discovery and data mining, KDD. ACM; New York, NY, USA: 2003. Empirical Bayesian data mining for discovering patterns in post-marketing drug safety; pp. 359–368. [Google Scholar]
- 37.Almenoff J.S., LaCroix K.K., Yuen N.A., Fram D., DuMouchel W. Comparative performance of two quantitative safety signalling methods. Drug Saf. 2006;29(10):875–887. doi: 10.2165/00002018-200629100-00005. [DOI] [PubMed] [Google Scholar]
- 38.DuMouchel W., Harpaz R. Regression-adjusted GPS algorithm (RGPS) Oracle Health Sci. 2015;(April) [Google Scholar]
- 39.Sequeira A., Mamdani F., Lalovic A., Anguelova M., Lesage A., Seguin M. Alpha 2A adrenergic receptor gene and suicide. Psychiatry Res. 2004;125(2):87–93. doi: 10.1016/j.psychres.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 40.Cottingham C., Wang Q. α2 adrenergic receptor dysregulation in depressive disorders: implications for the neurobiology of depression and antidepressant therapy. Neurosci Biobehav Rev. 2012;36(10):2214–2225. doi: 10.1016/j.neubiorev.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Curran M.E., Splawski I., Timothy K.W., Vincent G.M., Green E.D., Keating M.T. A molecular basis for cardiac arrhythmia: HERG mutations cause long QT syndrome. Cell. 1995;80(5):795–803. doi: 10.1016/0092-8674(95)90358-5. [DOI] [PubMed] [Google Scholar]
- 42.Wen D., Liu A., Chen F., Yang J., Dai R. Validation of visualized transgenic zebrafish as a high throughput model to assay bradycardia related cardio toxicity risk candidates. J Appl Toxicol. 2012;32(10):834–842. doi: 10.1002/jat.2755. [DOI] [PubMed] [Google Scholar]
- 43.Mitcheson J.S. hERG potassium channels and the structural basis of drug-induced arrhythmias. Chem Res Toxicol. 2008;21(5):1005–1010. doi: 10.1021/tx800035b. [DOI] [PubMed] [Google Scholar]
- 44.Yusof I., Shah F., Hashimoto T., Segall M.D., Greene N. Finding the rules for successful drug optimisation. Drug Discov Today. 2014;19(5):680–687. doi: 10.1016/j.drudis.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 45.Movsesian M., Wever-Pinzon O., Vandeput F. PDE3 inhibition in dilated cardiomyopathy. Curr Opin Pharmacol. 2011;11(6):707–713. doi: 10.1016/j.coph.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Knight W., Yan C. Therapeutic potential of PDE modulation in treating heart disease. Future Med Chem. 2013;5(14):1607–1620. doi: 10.4155/fmc.13.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ely S.W., Matherne G.P., Coleman S.D., Berne R.M. Inhibition of adenosine metabolism increases myocardial interstitial adenosine concentrations and coronary flow. J Mol Cell Cardiol. 1992;24(11):1321–1332. doi: 10.1016/0022-2828(92)93097-4. [DOI] [PubMed] [Google Scholar]
- 48.Aviña-Zubieta J.A., Abrahamowicz M., De Vera M.A., Choi H.K., Sayre E.C., Rahman M.M. Immediate and past cumulative effects of oral glucocorticoids on the risk of acute myocardial infarction in rheumatoid arthritis: a population-based study. Rheumatology. 2013;52(1):68–75. doi: 10.1093/rheumatology/kes353. [DOI] [PubMed] [Google Scholar]
- 49.Oakley R.H., Cidlowski J.A. Glucocorticoid signaling in the heart: a cardiomyocyte perspective. J Steroid Biochem Mol Biol. 2015;153:27–34. doi: 10.1016/j.jsbmb.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huerta C., Castellsague J., Varas-Lorenzo C., García Rodríguez L.A. Nonsteroidal anti-inflammatory drugs and risk of ARF in the general population. Am J Kidney Dis. 2005;45(3):531–539. doi: 10.1053/j.ajkd.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 51.Wang L., Sha Y., Bai J., Eisner W., Sparks M.A., Buckley A.F. Podocyte-specific knockout of cyclooxygenase 2 exacerbates diabetic kidney disease. Am J Physiol Renal Physiol. 2017;313(2):F430–F439. doi: 10.1152/ajprenal.00614.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Slattery P., Frölich S., Schreiber Y., Nüsing R.M. COX-2 gene dosage-dependent defects in kidney development. Am J Physiol Renal Physiol. 2016;310(10):F1113–F1122. doi: 10.1152/ajprenal.00430.2015. [DOI] [PubMed] [Google Scholar]
- 53.Ye H., Wang X., Sussman C.R., Hopp K., Irazabal M.V., Bakeberg J.L. Modulation of polycystic kidney disease severity by phosphodiesterase 1 and 3 subfamilies. J Am Soc Nephrol. 2016;27(5):1312–1320. doi: 10.1681/ASN.2015010057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wadhwa S., Wadhwa P., Dinda A.K., Gupta N.P. Differential expression of potassium ion channels in human renal cell carcinoma. Int Urol Nephrol. 2009;41(2):251–257. doi: 10.1007/s11255-008-9459-z. [DOI] [PubMed] [Google Scholar]
- 55.Fagerberg L., Hallström B.M., Oksvold P., Kampf C., Djureinovic D., Odeberg J. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13(2):397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nussey S.S., Whitehead S.A. CRC Press; 2013. Endocrinology: an integrated approach. [PubMed] [Google Scholar]
- 57.Miyamoto J., Matsumoto T., Shiina H., Inoue K., Takada I., Ito S. The pituitary function of androgen receptor constitutes a glucocorticoid production circuit. Mol Cell Biol. 2007;27(13):4807–4814. doi: 10.1128/MCB.02039-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nuzzi R., Scalabrin S., Becco A., Panzica G. Gonadal hormones and retinal disorders: a review. Front Endocrinol. 2018;9:66. doi: 10.3389/fendo.2018.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ashton H. Guidelines for the rational use of benzodiazepines. Drugs. 1994;48(1):25–40. doi: 10.2165/00003495-199448010-00004. [DOI] [PubMed] [Google Scholar]
- 60.Kubitz R., Dröge C., Stindt J., Weissenberger K., Häussinger D. The bile salt export pump (BSEP) in health and disease. Clin Res Hepatol Gastroenterol. 2012;36(6):536–553. doi: 10.1016/j.clinre.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 61.Riede J., Poller B., Huwyler J., Camenisch G. Assessing the risk of drug-induced cholestasis using unbound intrahepatic concentrations. Drug Metab Dispos. 2017;45(5):523–531. doi: 10.1124/dmd.116.074179. [DOI] [PubMed] [Google Scholar]
- 62.Liu Ll-Y, Wang X.-.H., Lu Y., Zhu Q.-.R., Wang J.-.S. Association of variants of ABCB11 with transient neonatal cholestasis : ABCB11 and TNC. Pediatr Int. 2013;55(2):138–144. doi: 10.1111/ped.12049. [DOI] [PubMed] [Google Scholar]
- 63.Geenes V., Williamson C. Intrahepatic cholestasis of pregnancy. World J Gastroenterol. 2009;15(17):2049. doi: 10.3748/wjg.15.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu M., Wu Y., Chen Y., Sun J., Zhao Z., Chen X.-.W. Large-scale prediction of adverse drug reactions using chemical, biological, and phenotypic properties of drugs. J Am Med Inform Assoc. 2012;19(e1):e28–e35. doi: 10.1136/amiajnl-2011-000699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bender A., Scheiber J., Glick M., Davies J.W., Azzaoui K., Hamon J. Analysis of pharmacology data and the prediction of adverse drug reactions and off-target effects from chemical structure. ChemMedChem: Chem Enab Drug Discov. 2007;2(6):861–873. doi: 10.1002/cmdc.200700026. [DOI] [PubMed] [Google Scholar]
- 66.Cheng J., Grande J.P. Cyclic nucleotide phosphodiesterase (PDE) inhibitors: novel therapeutic agents for progressive renal disease. Exp Biol Med. 2007;232(1):38–51. [PubMed] [Google Scholar]
- 67.Dawson S., Stahl S., Paul N., Barber J., Kenna J.G. In vitro inhibition of the bile salt export pump correlates with risk of cholestatic drug-induced liver injury in humans. Drug Metab Dispos. 2012;40(1):130–138. doi: 10.1124/dmd.111.040758. [DOI] [PubMed] [Google Scholar]
- 68.Montanari F., Pinto M., Khunweeraphong N., Wlcek K., Sohail M.I., Noeske T. Flagging drugs that inhibit the bile salt export pump. Mol Pharm. 2016;13(1):163–171. doi: 10.1021/acs.molpharmaceut.5b00594. [DOI] [PubMed] [Google Scholar]
- 69.Breiman L. Random forests. Mach Learn. 2001;45(1):5–32. [Google Scholar]
- 70.Charte F., Charte D. Working with multilabel datasets in R: the mldr package. The R Journal. 2015;7(2):149–162. [Google Scholar]
- 71.Rivolli A., de Carvalho A.C. The utiml package: multi-label classification in R. The R Journal. 2018;10(2):24–37. [Google Scholar]
- 72.Saito T., Rehmsmeier M. Precrec: fast and accurate precision–recall and ROC curve calculations in R. Bioinformatics. 2017;33(1):145–147. doi: 10.1093/bioinformatics/btw570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grau J., Grosse I., Keilwagen J. PRROC: computing and visualizing precision-recall and receiver operating characteristic curves in R. Bioinformatics. 2015;31(15):2595–2597. doi: 10.1093/bioinformatics/btv153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Menze B.H., Kelm B.M., Masuch R., Himmelreich U., Bachert P., Petrich W. A comparison of random forest and its Gini importance with standard chemometric methods for the feature selection and classification of spectral data. BMC Bioinformatics. 2009;10:213. doi: 10.1186/1471-2105-10-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brugada R., Hong K., Dumaine R., Cordeiro J., Gaita F., Borggrefe M. Sudden death associated with short-QT syndrome linked to mutations in HERG. Circulation. 2004;109(1):30–35. doi: 10.1161/01.CIR.0000109482.92774.3A. [DOI] [PubMed] [Google Scholar]
- 76.Pinto C.S., Raman A., Reif G.A., Magenheimer B.S., White C., Calvet J.P. Phosphodiesterase isoform regulation of cell proliferation and fluid secretion in autosomal dominant polycystic kidney disease. J Am Soc Nephrol. 2016;27(4):1124–1134. doi: 10.1681/ASN.2015010047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang X., Ward C.J., Harris P.C., Torres V.E. Cyclic nucleotide signaling in polycystic kidney disease. Kidney Int. 2010;77(2):129–140. doi: 10.1038/ki.2009.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Babcock J.J., Li M. hERG channel function: beyond long QT. Acta Pharmacol Sin. 2013;34(3):329–335. doi: 10.1038/aps.2013.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mertens K.L., Kalsbeek A., Soeters M.R., Eggink H.M. Bile acid signaling pathways from the enterohepatic circulation to the central nervous system. Front Neurosci. 2017;11:617. doi: 10.3389/fnins.2017.00617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Srivastava A. Progressive familial intrahepatic cholestasis. J Clin Exp Hepatol. 2014;4(1):25–36. doi: 10.1016/j.jceh.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Strautnieks S.S., Bull L.N., Knisely A.S., Kocoshis S.A., Dahl N., Arnell H. A gene encoding a liver-specific ABC transporter is mutated in progressive familial intrahepatic cholestasis. Nat Genet. 1998;20(3):233–238. doi: 10.1038/3034. [DOI] [PubMed] [Google Scholar]
- 82.Watanabe M., Houten S.M., Mataki C., Christoffolete M.A., Kim B.W., Sato H. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439(7075):484–489. doi: 10.1038/nature04330. [DOI] [PubMed] [Google Scholar]
- 83.Mukaisho K.-.I., Araki Y., Sugihara H., Tanaka H., Chen K.-.H., Hattori T. High serum bile acids cause hyperthyroidism and goiter. Dig Dis Sci. 2008;53(5):1411–1416. doi: 10.1007/s10620-007-0017-9. [DOI] [PubMed] [Google Scholar]
- 84.Comhair S.A.A., McDunn J., Bennett C., Fettig J., Erzurum S.C., Kalhan S.C. Metabolomic endotype of asthma. J Immunol. 2015;195(2):643–650. doi: 10.4049/jimmunol.1500736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tan X., Gao F., Su H., Gong Y., Zhang J., Sullivan M.A. Genetic and proteomic characterization of bile salt export pump (BSEP) in snake liver. Sci Rep. 2017;7:43556. doi: 10.1038/srep43556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li C.-.H., Sung F.-.C., Wang Y.-.C., Lin D., Kao C.-.H. Gallstones increase the risk of developing renal stones: a nationwide population-based retrospective cohort study. QJM. 2014;107(6):451–457. doi: 10.1093/qjmed/hcu017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is made available in Supplementary Tables 1–11 and on GitHub (https://github.com/samanfrm/ADRtarget).