Figure 2.
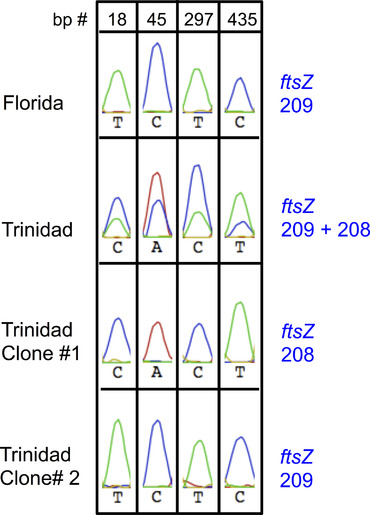
Examples of sequencing chromatograms illustrating strong Wolbachia “coinfection patterns” detected in this study and how clone library sequencing was used to validate these observations. The origins of the samples and the corresponding allele numbers (based on the multilocus sequence typing [MLST] database) are shown on the left and right, respectively. Strong coinfection patterns can be detected at various nucleotides of specific gene sequences (in this case at bp 18, 45, 297, and 435 of a ftsZ fragment). A sample originated from Lake Alfred, Florida only carries allele 209, while a coinfected sample collected from Trinidad contains more than one allele. Screening of clone libraries constructed from the Trinidad sample identified clones carrying different alleles (alleles 208 and 209; lower two panels) and explained the patterns seen in the original chromatogram. The peaks shown are parts of the actual chromatograms of the sequencing data (in the Geneious software). Peaks with different colors represent different nucleotides. Red: A; Green: T; Blue: C.
