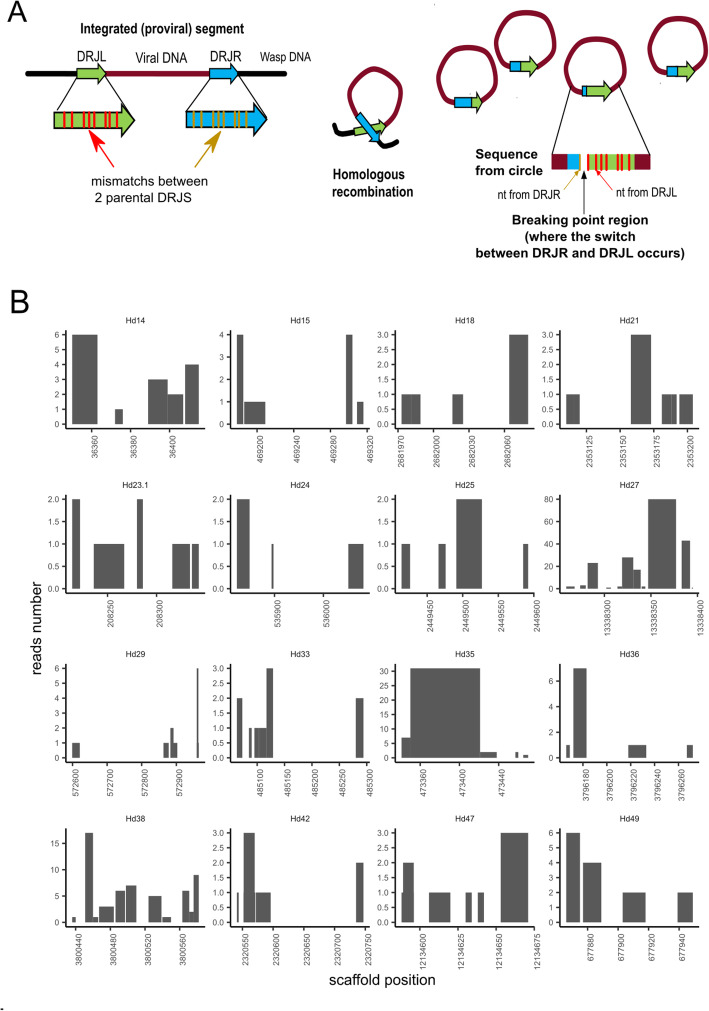
Fig. 3.
Segment DRJ variability in terms of excision sites in Hyposoter didymator. a Schematic representation of the homologous recombination between the two DRJs flanking the proviral sequence (left DRJL and right DRJR ends) to produce the circular molecule (segment) containing one recombined DRJ sequence. When excision sites are located at different positions in the DRJ, segments differing in their recombined DRJ sequence are generated. Excision occurs more frequently at some positions, resulting in different relative amounts of each isoform. On the right, rationale of the algorithm developed to identify the “break points.” Mapping of the segment sequence (DRJsegment) with the two parental DRJs, which differ in their sequences (nucleotide (nt) mismatches), allows identification of the regions where the switch from one parental DRJ to the other has occurred (in the diagram, between the first and second mismatch). b Prediction of putative recombination break points in H. didymator DRJs. Each graph corresponds to the left copy of the DRJ for a given segment (indicated below each graph). The X-axis is the position in the scaffold. The Y-axis indicates the number of reads (obtained from sequencing of the packaged circular DNA molecules) confirming that the circle has been recombined between these two positions, based on the observed mismatches at both end of the segment for each read. We observed between 1 and 80 reads per breakpoint region according to the analyzed segment