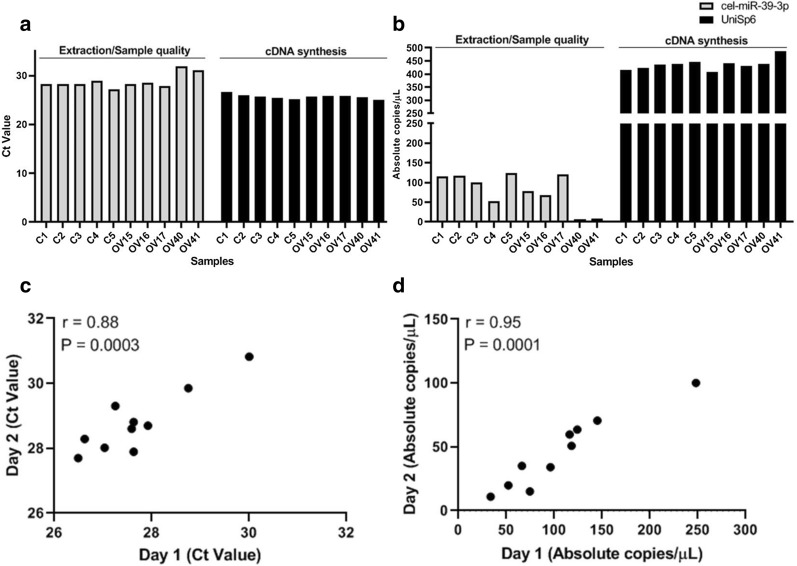
Fig. 1.
Optimization of cel-miR-39-3p and UniSp6 spike-ins and assay reproducibility for circulating miR-320a detection. All the 10 serum samples (5 from health-donors and 5 from ovarian cancer patients) were spiked-in with 0.2 nM of cel-miR-39-3p (gray bars) during extraction step and 0.2 µL of UniSp6 (black bars) were add during the cDNA synthesis. a RT-qPCR and b ddPCR were used to quantify the levels of both miRNAs. For the circulating miR-320a quantification, Pearson correlation (r) was applied to 2 independent datasets for all 10 samples to calculate the reproducibility of detection of miR-320a by (c) RT-qPCR and (d) ddPCR. “Day 1” and “Day 2” means two independent experiment (2 independent batches). Ct cycle threshold, C Health-controls, OV ovarian cancer patients