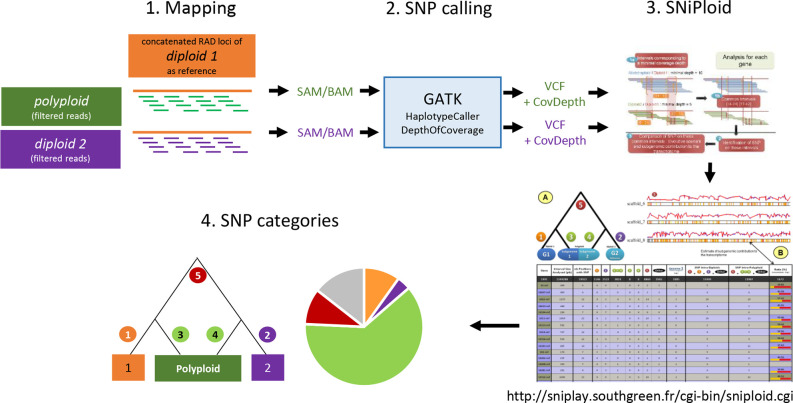
Figure 1.
SNiPloid workflow adapted from (Peralta et al., 2013). The filtered RAD sequencing reads of the polyploid and one putative parental species (Diploid 2) are mapped to the concatenated RAD loci of the second putative parental species (Diploid 1) that act as a “pseudo-reference”. SNPs and coverage depth are determined with GATK tools “HaplotypeCaller” and “DepthOfCoverage”. The resulting “VCF” and “CovDepth” files of both samples serve as input for SNiPloid. The output of SNiPloid contain the assigned SNP categories 1‑5 for each compared SNP. Those data are finally summarized in a pie chart that shows the proportions of the observed categories.