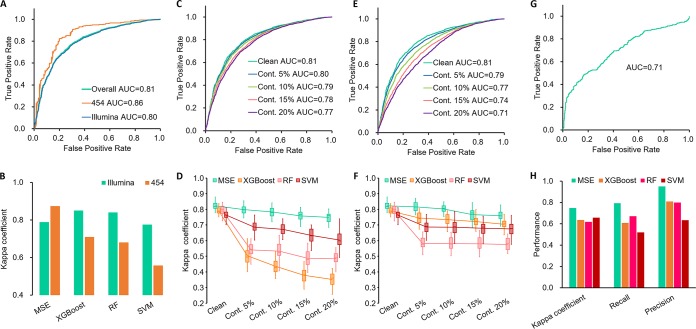
FIG 3.
Robustness of MSE to sequencing platform change and DNA contamination. Gut microbiome samples were used as the example. (A) ROCs of MNS-based disease status prediction under two sequencing platforms. (B) Difference in Kappa coefficient (k) of disease classification under two sequencing platforms. ROCs of MNS (C) and variation of k for multiple-disease classification (D) with reagent blank microbiome contaminations. ROCs of MNS (E) and variation of k of multiple disease classification (F) with indoor environmental contaminations. (G) ROC of MNS-based disease detection by independent IBD cohorts and (H) disease classification for cross-cohort data sets by MSE and by model-based approaches. For boxplots in D and F, central lines represent the medians, the bounds of the box represent the quartiles, and error bars represent the local maximum and local minimum values. Source data are provided as Data Set S1.