FIGURE 2.
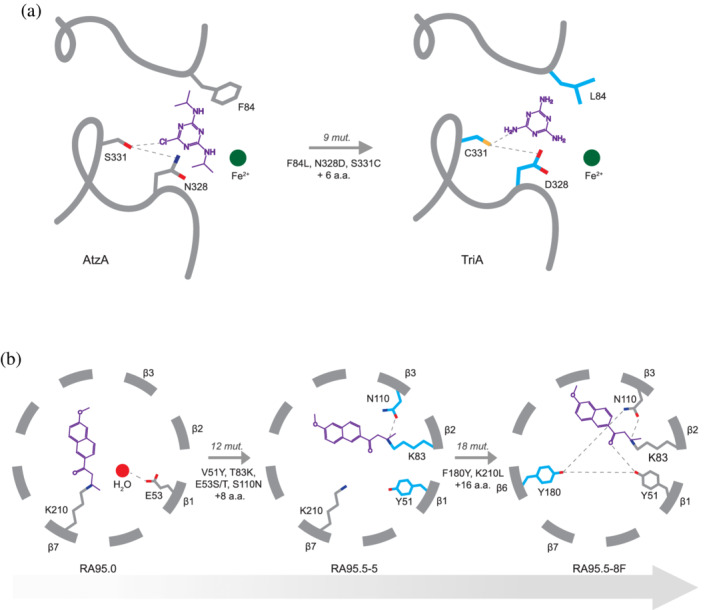
Creation of new enzyme–substrate interactions by evolution. (a) Schematic representation of key active site residues in AtzA and TriA. The Ser–Asn dyad, involved in atrazine dechlorination, is substituted to Cys–Asp during the evolution towards melamine deamination. 18 An additional active site residue, Phe84, is also mutated to leucine. The active site Fe2+ is depicted as a green sphere. (b) Schematic representation of the stepwise evolution of a designed retro‐aldolase, RA95.0. 55 , 57 (left) The catalytic dyad (K210‐E53 and a water molecule) from the initial RA95.0 design, (centre) was mutated to a distinct triad (K83‐N110‐Y51) in RA95.5–5 during directed evolution. (right) Further evolution resulted in the emergence of a catalytic tetrad (K83‐N110‐Y51‐Y180) in RA95.5‐8F. Key active site residues for Panels A and B are depicted as sticks; new residues installed at each evolutionary stage are highlighted in blue. The mutations fixed between each step of the trajectory are indicated on the gray arrows. The chemical structures of atrazine (left) and melamine (right) in Panel A, and the 1,3‐diketone mechanism‐based inhibitor of retro‐aldolases in Panel B are shown in purple. Panel B is adapted from Ref. 55
