FIGURE 5.
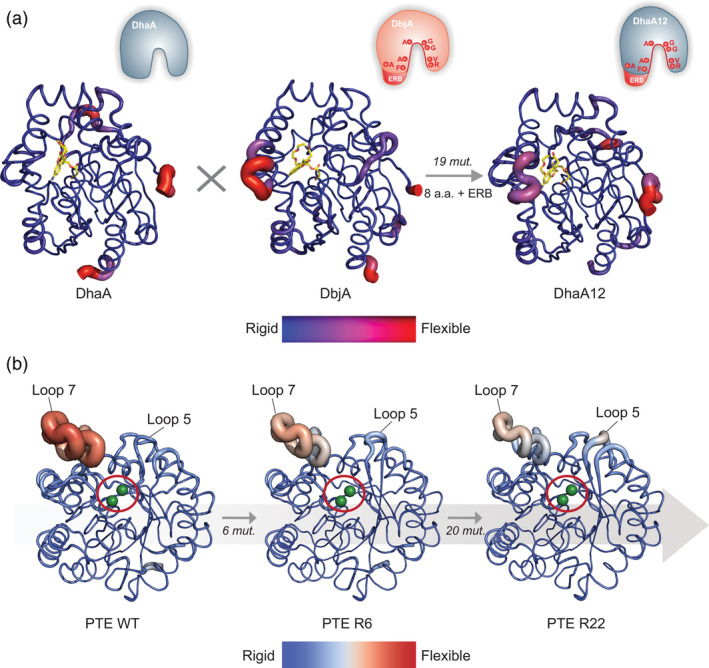
Alteration of enzyme conformational dynamics by evolution. Backbone B‐factors of (a) DhaA, DbjA, and DhaA12 haloalkane dehydrogenases, obtained by molecular dynamics simulations (MDs), 27 and (b) phosphotriesterase (PTE) WT, and variants R6, and R22 (PDB ID: 4pcp, 4xag, 4pcn) crystal structures, shown as cartoon putty representation. 46 The B‐factors are visualized by a color scale mapped onto the structure, and ribbon thickness. (a) The average structures of covalently bound fluorescent probes are shown as yellow sticks. A schematic of the active site transplantation events (mutations and ERB) is represented in the upper right corner of each enzyme. (b) The active site location of PTE is indicated by a red circle, and two Zn2+ ions shown as green spheres. The number of mutations fixed between each step of the trajectory is indicated on the gray arrow. Panel A is adapted with permission from Ref. 27
