FIGURE 6.
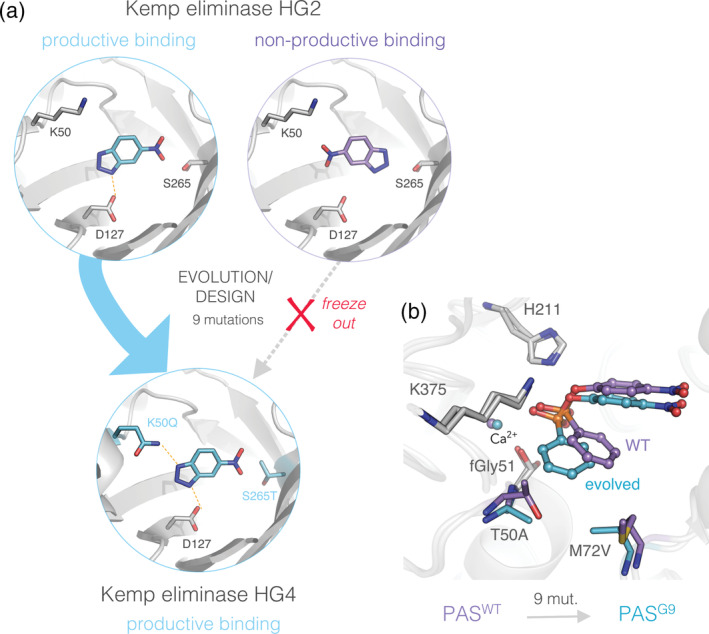
Substrate repositioning leads to novel enzyme‐substrate complexes in evolution. (a) The evolution of a designed kemp eliminase produced a weakly active mutant, HG2 (PDB ID: 3nyd), exhibiting two binding modes for the TS analog, 6‐nitrobenzotriazole. 108 (left) Asp127 binds the substrate in a productive orientation (teal sticks), as designed. (right) However, a nonproductive, flipped orientation (purple sticks), is also observed in the crystal structure. Further evolution stabilized the productive complex upon mutations K50Q and S265T, which eliminated the nonproductive orientation in subsequent mutants, for example, in the most evolved HG4 variant (PDB ID: 5rgf). 51 , 105 (b) Substrate repositioning induced by mutations T50A and M72V, from PASWT (PDB ID: 1hdh) to PASG9 (PDB ID: 4cxk) during arylsulfatase (PAS) evolution toward phosphonate monoester hydrolysis. 29 Evolution shifted the substrate closer to the catalytic machinery (Fgly nucleophile, K375 and H211) without altering the position of the catalytic residues, or creating new enzyme‐substrate interactions. Snapshots are representative stationary points from molecular dynamics simulations (MDs)
