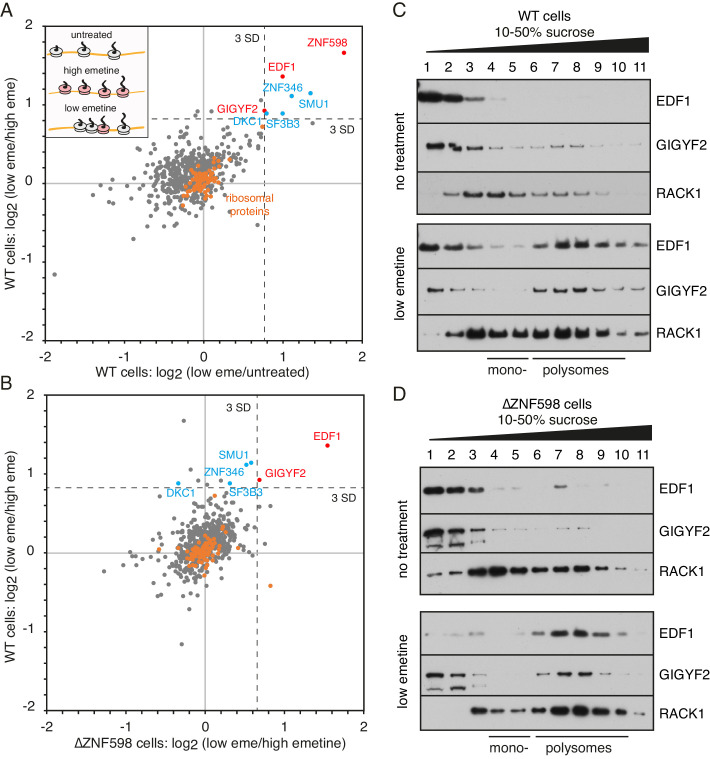
Figure 1. Proteomic identification of collision-specific ribosome interacting factors.
(A) Plot of proteins identified by quantitative mass spectrometry in the polysome fraction of cytosol separated by sucrose gradient centrifugation. The cells were either left untreated, treated with low-dose emetine (1.8 µM) to induce collisions (low eme), or treated with high-dose emetine (360 µM) to freeze ribosomes in place (high eme). Pairwise comparisons are plotted such that factors specifically associating with collided ribosomes (i.e., only in low-dose emetine samples) should fall in the upper-right quadrant. Ribosomal proteins are indicated in orange. The position of 3 standard deviations (SD) from the mean along each axis is indicated with a dashed line. Proteins that deviate 3 SD or more along both axes are labelled (in blue and red). (B) Cells knocked out for ZNF598 (∆ZNF598) were treated with low or high dose emetine as in panel A and the polysome fractions were analysed by mass spectrometry. These data (x-axis) are plotted relative to the same comparison in wild type cells (y-axis). The position of 3 SD along each axis is indicated with a dashed line, and the data points for the same proteins from panel A are labelled. Note that only EDF1 and GIGYF2 fall into the upper right quadrant in both plots. As expected, ZNF598 was not detected in ∆ZNF598 cells. (C, D) Sucrose gradient fractionation and immunoblots of cytosolic lysates prepared from untreated cells versus low-dose emetine treated cells. WT cells were analysed in panel C and ∆ZNF598 cells in panel D. RACK1 is a 40S-ribosomal protein. The positions of fractions containing mono- and polysomes are indicated. Source data are provided for the mass spectrometry analysis.