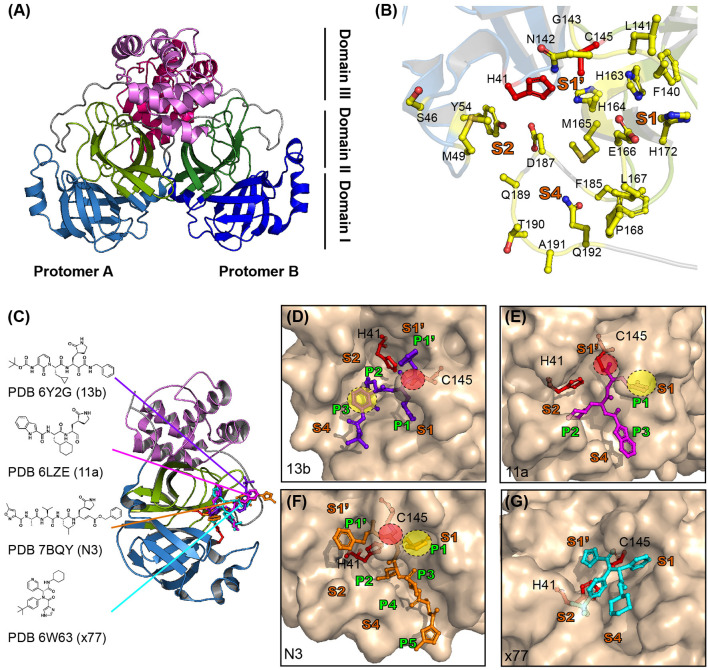
Figure 3.
Structure of SARS-CoV-2 viral Mpro and its complex with inhibitors. (A) The crystal structure of SARS-CoV-2 Mpro. Mpro is a cysteine protease that consists of three domains and two protomers. Protomer B is shown in darker colors than protomer A and each domain is shown in different colors (sky blue, split pea, and violet represent domains 1, 2, and 3, respectively). (B) Substrate binding site of SARS-CoV-2 Mpro. The substrate binding site of Mpro is subdivided into S1, S1′, S2, and S4 (shown in bold orange). The inhibitors bind to 17 residues shown as yellow sticks (H41, S46, M49, Y56, F140, L141, N142, C145, H164, M165, E166, L167, H172, Q189, F185, T190, and Q192). The cysteine-histidine dyad (C145-H41) between domains 1 and 2 is shown in red. (C) SARS-CoV-2 Mpro in complex with its inhibitors. The structures of SARS-CoV-2 Mpro in complex with 13b (PDB ID 6Y2G, purple sticks), 11a (PDB ID 6LZ2, magenta sticks), N3 (PDB ID 7BQY, orange sticks), and x77 (PDB ID 6W63, cyan sticks) are shown. The molecular interaction of each inhibitor in the active site is shown as a surface and stick complex (D–G are 13b, 11a, N3, and x77). The γ-lactam ring that plays an important inhibitory role is shown in the yellow circle, and C-S covalent bonds with Cys145 are shown in the red circle.