Fig. 1 |. DddA is a double-stranded DNA cytidine deaminase that mediates T6SS-dependent interbacterial antagonism.
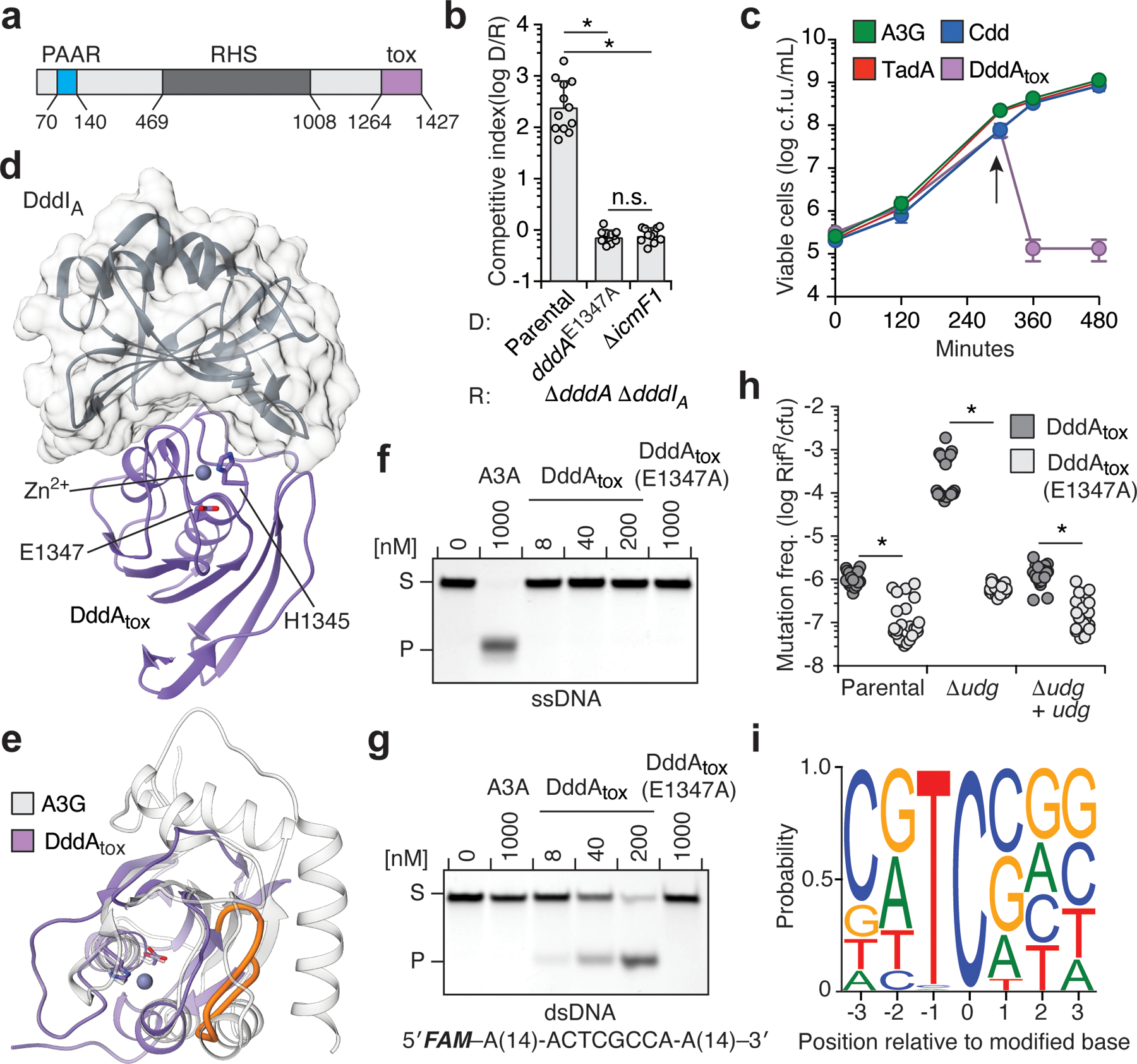
a, Domains of full-length DddA. PAAR, proline-alanine-alanine-arginine; RHS, rearrangement hotspot; Tox, toxin domain. b, Competitiveness of the indicated donor B. cenocepacia strains (D) towards the B. cenocepacia ΔdddAΔdddIA recipient strain (R). c, Viability of E. coli populations expressing the indicated deaminases, induced at 300 min (arrow). A3G, APOBEC3G; Cdd, E. coli cytidine deaminase; TadA, tRNA adenosine deaminase A; cfu, colony-forming units. d, Crystal structure of DddAtox (purple) complexed with DddIA (grey). e, Structural alignment of DddAtox (purple) and APOBEC3G (white). The intervening loop of DddAtox that is absent in APOBEC3G is shown in orange. f, g, In vitro cytidine deamination assays using a single-stranded (f) or double-stranded (g) 36-nt 6-carboxyfluorescein (FAM)-labelled DNA substrate (S), which contains AC, TC, CC and GC as indicated in g. Cytidine deamination leads to products (P) with increased mobility. A3A, APOBEC3A. Gels are representative of three replicates. h, Mutation frequency in E. coli strains expressing DddAtox or catalytically inactive DddAtox(E1347A). pBAD24::udg was used for complementation of Δudg (+udg). Values are derived from eight independent biological replicates. RifR, rifampicin resistant colonies. i, Probability sequence logo of the region flanking mutated cytosines in five E. coli Δudg isolates serially exposed to a low level of DddAtox. Values and error bars reflect mean ± s.d. of n = 4 (in b) or n = 3 (in c) independent biological replicates. *P < 0.0001; NS, not significant (P > 0.05) by Student’s unpaired two-tailed t-test.
