Extended Data Fig. 5 |. Unoptimized mitoTALE–split DddAtox fusions mediate modest editing of mitochondrial ND6 in HEK293T cells.
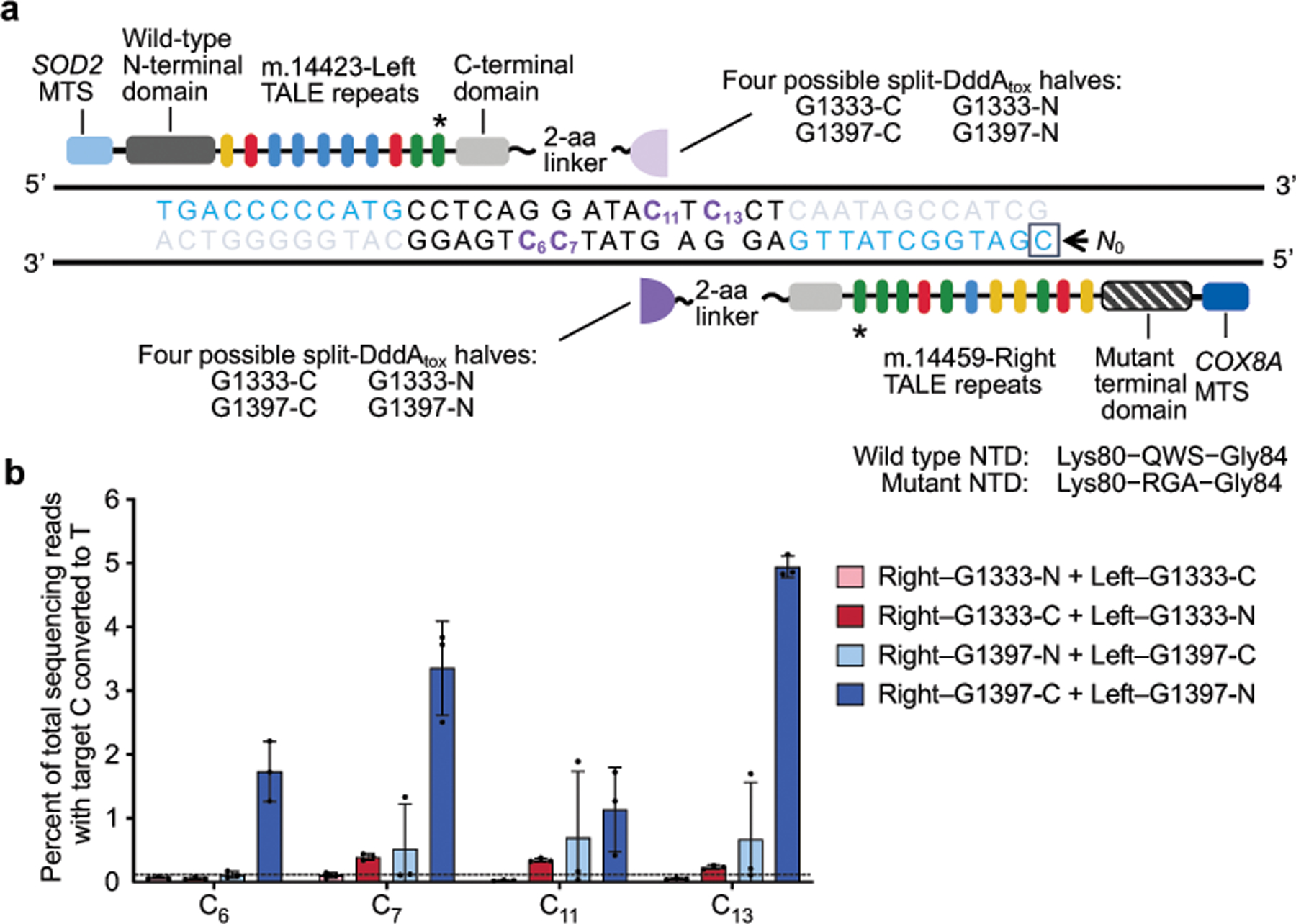
a, Architectures of non-UGI containing ND6-mitoTALE–DddAtox fusion pair. DddAtox was split at G1333 or G1397, with each half fused to either the left TALE or the right TALE. TALEs bind to mtDNA sequences (blue) that flank a 15-bp spacing region in mitochondrial ND6. Target cytosines are shown in purple. The last TALE repeat (*) did not match the reference genome9 (see Supplementary Table 4). b, mtDNA editing efficiencies of mitoTALE–DddAtox pairs in the listed split orientations. The dashed line is drawn at 0.1%. Values and error bars reflect the mean ± s.d. of n = 3 independent biological replicates.
