Fig. 2 |. Non-toxic split-DddAtox halves reconstitute activity when co-localized on DNA in HEK293T cells.
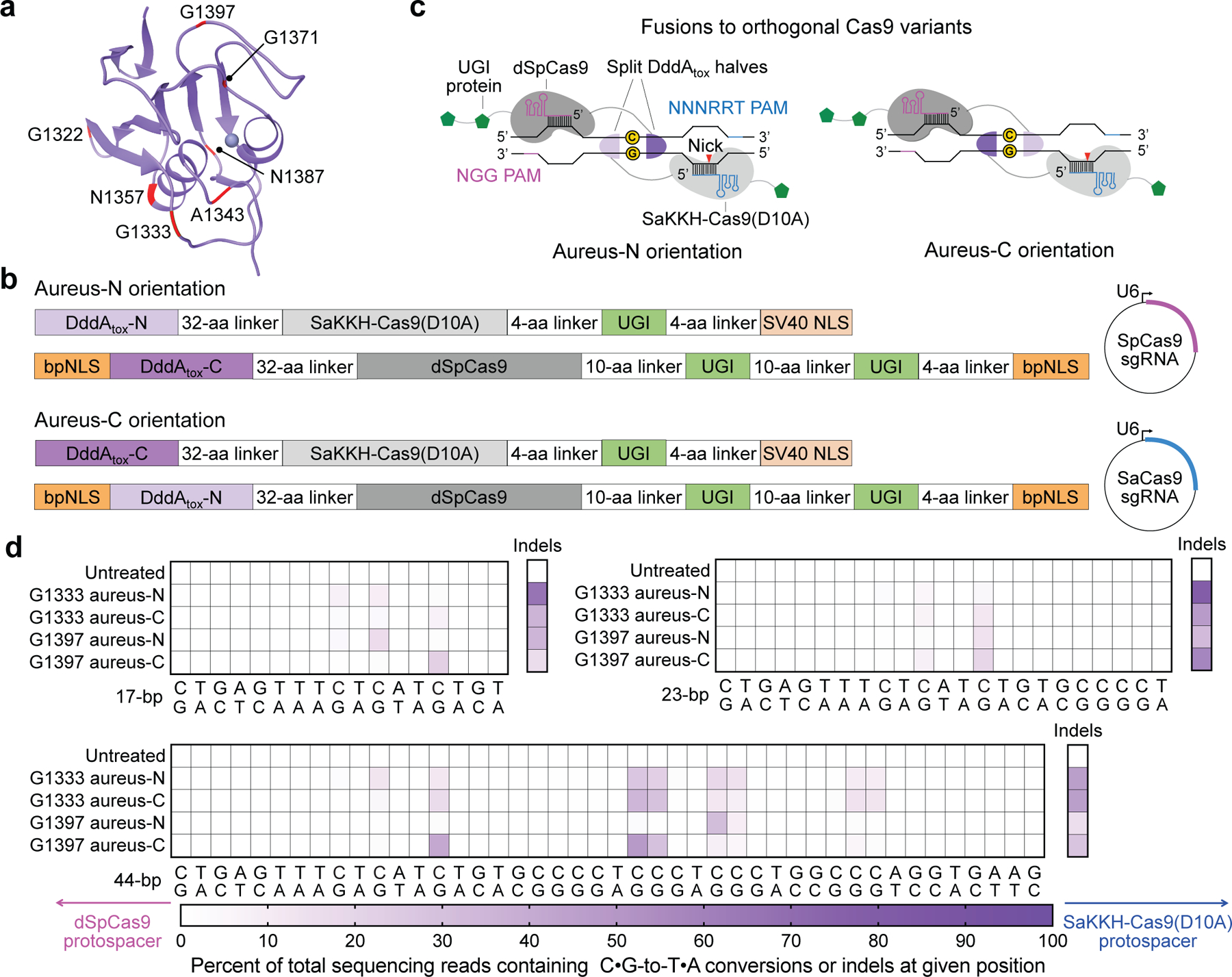
a, DddAtox was split at the peptide bond between each labelled amino acid and the following residue. b, Architectures of split-DddAtox–Cas9 fusions. DddAtox-N and DddAtox-C contain the N terminus and C terminus of DddAtox, respectively. Two fusion orientations (aureus-N or aureus-C) are possible for a given split. sgRNA, single guide RNA. c, Fusions of split-DddAtox halves to orthogonal Cas9 variants enable reassembly of active DddAtox, without creating non-functional homodimers. PAM, protospacer adjacent motif. d, Heat maps showing C•G-to-T•A conversion and indel frequencies for G1333 and G1397 splits at the nuclear EMX1. The split orientations and positions of dSpCas9 (pink) and SaKKH-Cas9(D10A) (blue) protospacers are shown. Colours reflect the mean of n = 2 independent biological replicates.
