Extended Data Fig. 1 |. Analysis of the bactericidal activity of DddA and its activity against dsDNA and RNA substrates.
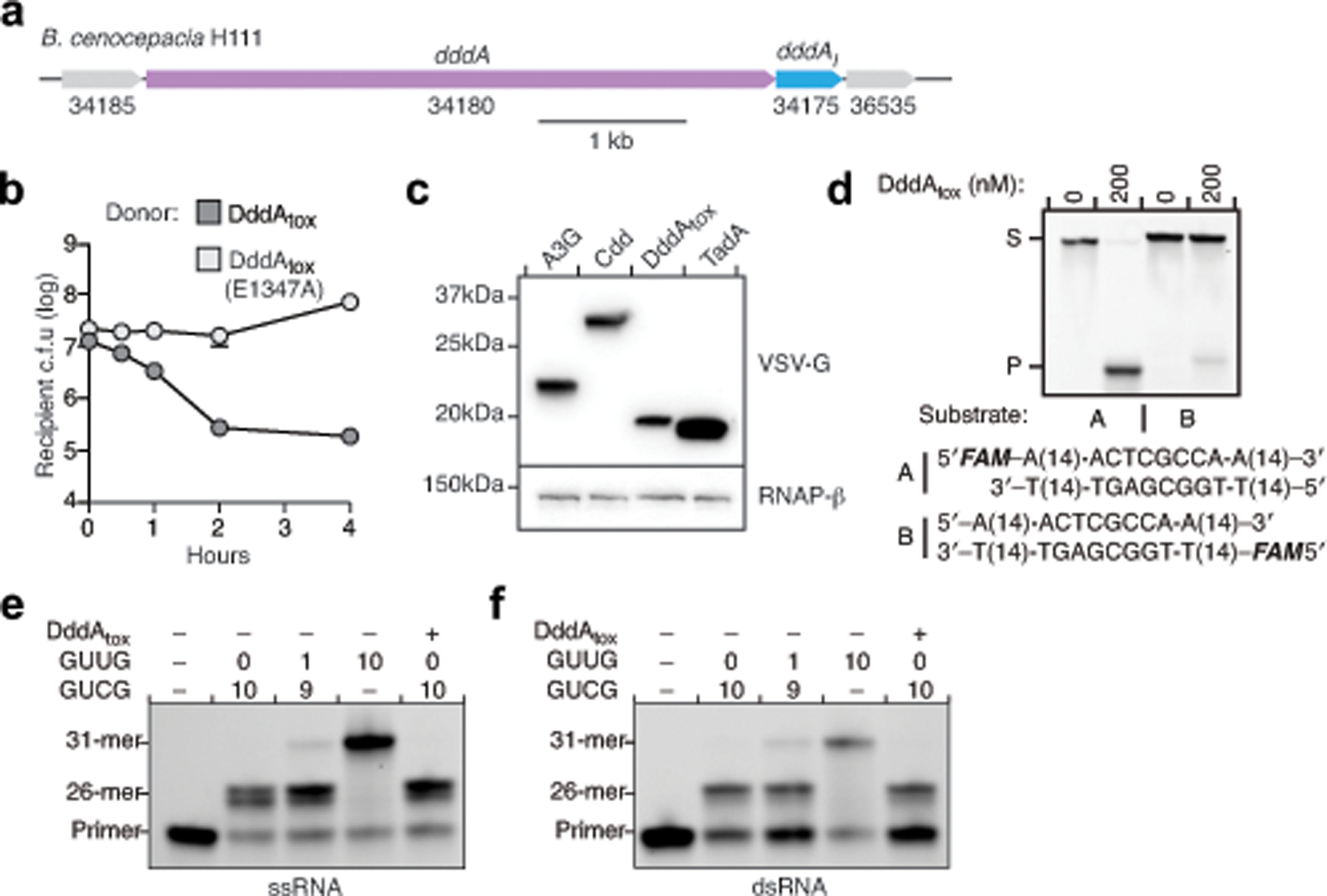
a, Genomic context of dddA (purple) and dddIA (blue) in B. cenocepacia H111. b, Viability of B. cenocepacia ΔdddA ΔdddIA (recipient) over time during competition with B. cenocepacia donor strains carrying wild-type dddAtox or dddAtoxE1347A. Values and error bars represent the mean ± s.d. of three technical replicates. The experiment was repeated three times with similar results. c, α-VSV-g western blot analysis of total cell lysates of E. coli expressing the indicated deaminases tagged with VSV-G epitope. RNAP-β was used as a loading control. Results are representative of n = 2 independent biological replicates. d, In vitro DNA cytidine deamination assays using double-stranded 36-nt DNA substrates containing AC, TC, CC, and GC with a FAM fluorophore on the forward (A) or reverse (B) strand. Deamination activity results in a cleaved product (P). Images are representative of n = 2 independent biological replicates. e, f, Poisoned primer extension assay to detect deamination of cytidine in single-stranded (e) or double-stranded (f) RNA substrates. Images are representative of n = 2 independent biological replicates. A mix of RNA substrates containing the sequences GUCG or GUUG at the indicated ratios were incubated with purified DddAtox and reverse transcriptase. Primer extension was performed in reactions with ddGTP to terminate primer extension at cytidines. Cytidine deamination yields the 31-mer product.
