Figure 3.
NEAR-Based COVID-19 Test
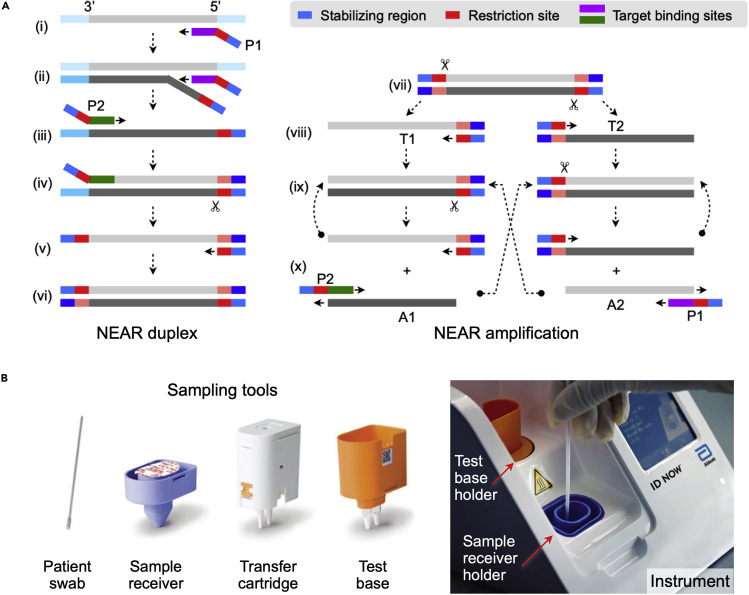
(A) Left, NEAR duplex formation. (i) The reverse primer (P1) binds to the target region and is extended. (ii) A second P1 binds to the same target and is extended, displacing the first extended strand. (iii) Primer P2 binds to the released strand and is extended, creating a double-stranded nicking enzyme recognition site. (iv) Nicking enzyme (indicated by scissors) binds and nicks the downstream of the recognition sequence. (v) Polymerase synthesizes complementary sequence off the cleaved site. (vi) The final product is a double-stranded DNA with restriction sites at both ends. Right, exponential amplification. (vii) Nicking enzyme binds and nicks the NEAR duplex at both restriction sides, making two templates (T1, T2). (viii) Free ends of templates are extended. (ix) Repeated nicking and polymerization steps start. (x) Cleaved complexes are regenerated, whereas amplified products (A1, A2) anneal to primers (P2, P1), resulting in bidirectional extension and creating duplexes.
(B) ID NOW COVID-19 system by Abbott. Disposable tools (left) minimize hands-on processes. Patient swab (nasal, nasopharyngeal, or throat) is eluted in the sample receiver containing elution/lysis buffer. After 10 s mixing, the mixture is manually transferred to the test base holder (via transfer cartridge) that contains lyophilized NEAR agents. Heating, agitation, and detection by fluorescence are performed automatically by the instrument. The assay detects SARS-CoV-2 RdRp gene. Adapted with permission from Ref (Nie et al., 2014).
Copyright 2014, American Society for Microbiology.