Figure 5.
CRISPR-Based COVID-19 Test
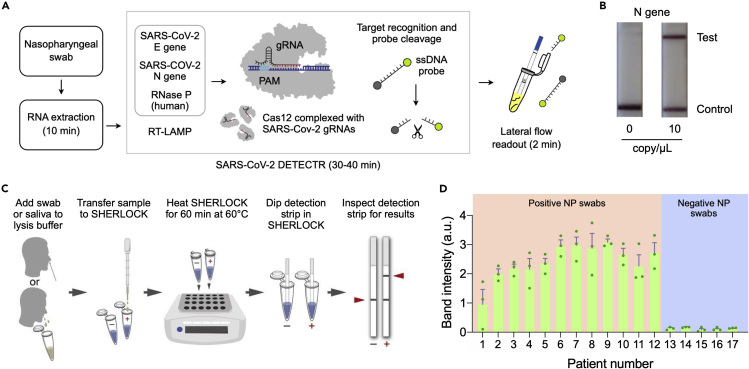
(A) Schematic of DETECTR coupled with lateral flow readout. RNA targets extracted from nasopharyngeal swabs are amplified by RT-LAMP. Cas12a complexes, pre-incubated with guide RNAs (gRNAs), recognize target DNA and cleave single-stranded DNA (ssDNA) probes for signal generation.
(B) The intact ddDNA reporters are captured on the control line, whereas the cleaved reporter is captured on the test line. Lateral flow results for the DETECTR are shown for N gene at 0 and 10 copy/μL.
(C) Schematic of SHERLOCK Testing in One Pot (STOP) test. A nasopharyngeal swab or saliva is transferred to the lysis buffer. Lysate is then added to SHERLOCK master mix, and the mixture is heated for 60 min at 60°C. Test results are read out using lateral flow strips (2 min).
(D) Twelve positive and five negative nasopharyngeal (NP) swab samples were analyzed by STOP. The assay made correct diagnosis of these samples. The data were displayed as mean ± standard deviation from three independent experiments.
(A and B) Adapted with permission from Ref (Broughton et al., 2020). Copyright 2020 Nature Publishing Group. (C and D) Adapted with permission from Ref (Joung et al., 2020).