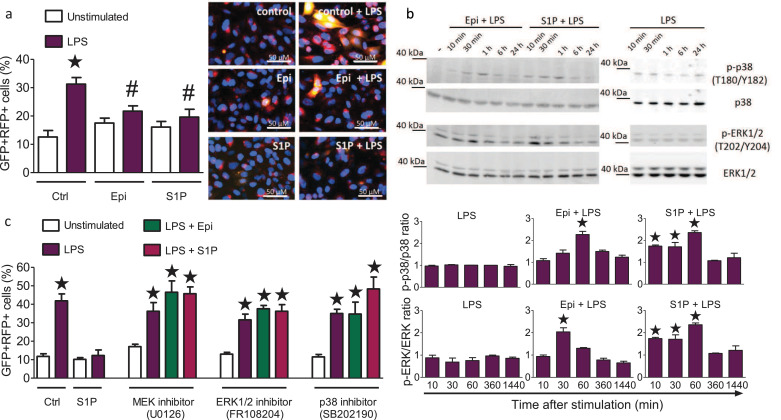
Fig. 4.
Epirubicin and S1P promote the reversal of LPS-induced autophagy via MAP kinase pathways.
(a) Autophagy in H1975 lung epithelial cells transduced with the Premo™ autophagy tandem sensor RFP-GFP-LC3B (30 particles/cell) after 24 h culture. Cells were treated with 200 ng/ml LPS in the absence or presence of 0.5 µM S1P or 0.6 ng/ml Epi. Representative cell images are shown, scale bars: 50 µm. The number of GFP and RFP double-positive cells per 100 cells was determined in 5 fields of 3 independent samples, means ± SEM, n = 5, *p<0.05 compared to untreated cells, #p<0.05 compared to LPS treated cells (One-way ANOVA with Bartlett´s test for equal variances and post-hoc Bonferroni's multiple comparison test). (b) Western blot detection and densitometric quantification of p38, ERK1/2, p-p38 and p-ERK1/2 in H1975 cells treated with 0.5 µM S1P or 0.6 ng/ml Epi along with 200 ng/ml LPS (left panel) or with 200 ng/ml LPS (right panel) for the indicated times, means ± SEM, n = 3, *p<0.05 (One-way ANOVA with Bartlett´s test for equal variances and post-hoc Bonferroni's multiple comparison test). (c) Autophagy in H1975 lung epithelial cells transduced with the Premo™ autophagy tandem sensor RFP-GFP-LC3B (30 particles/cell). Cells were stimulated for 24 h without and with LPS and treated with 10 µM U0126 (MEK inhibitor), 50 µM FR108204 (ERK1/2 inhibitor), or 10 µM SB201190 (p38 inhibitor) without and with S1P (0.5 µM) or Epi (0.6 ng/ml) as indicated. The number of both GFP and RFP double-positive cells per 100 cells were counted in 5 fields of 3 wells per stimulation, means ± SEM, n = 3, *p<0.05 (One-way ANOVA with Bartlett´s test for equal variances and post-hoc Bonferroni's multiple comparison test).