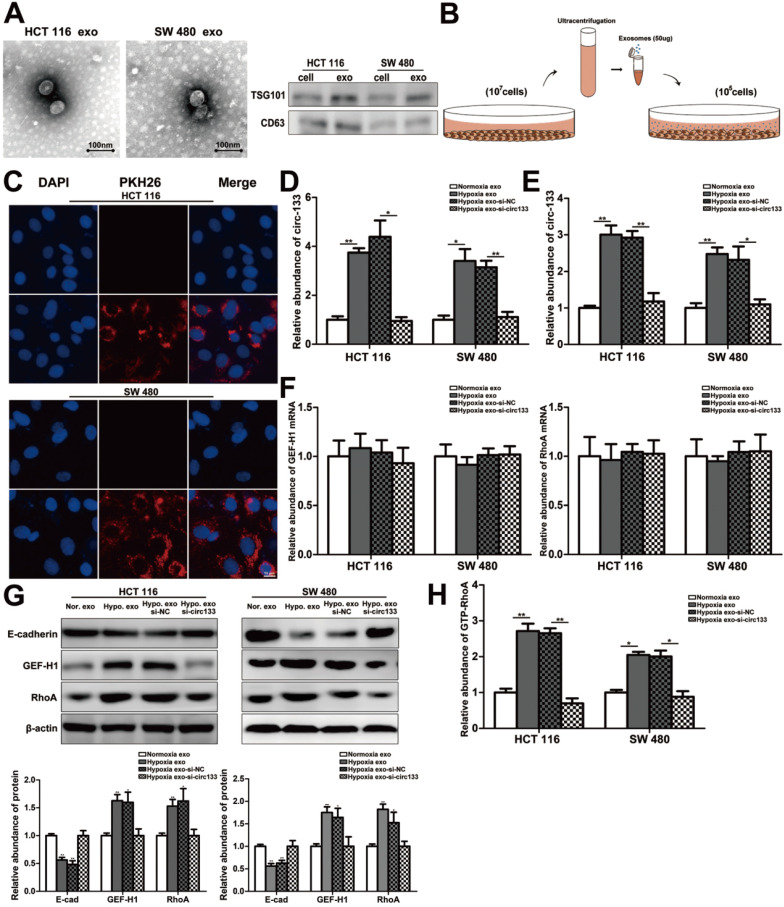
Figure 4.
Hypoxic exosomes incubate normoxic CRC cells. (A) Electron microscope scanning of exosomes derived from HCT116 and SW480 cells. And western blot analysis of exosome-enriched protein CD63 and key proteins for miRNA function, TSG101 (n=3). (B) Schematic description of the experimental design. (C) Confocal microscopy image of the internalization of fluorescently labelled exosomes in HCT116 and SW480 cells. (D) Quantitative RT-PCR analysis of the content of circ-133 in normoxia exosome, hypoxia exosome, hypoxia si-NC exosomes and hypoxia si-circ-133 exosomes, (n=3). (E)The content of circ-133 in HCT116 and SW480 cells that pretreated with normoxia exosome, hypoxia exosome, hypoxia si-NC exosomes and hypoxia si-circ-133 exosomes, respectively, (n=3). (F) The level of GEF-H1 mRNA and RhoA mRNA in each treatment group (treated with normoxia exosome, hypoxia exosome, hypoxia si-NC exosomes and hypoxia si-circ-133 exosomes, respectively), (n=3). (G) Associated proteins expression in each treatment groups, (n=3). (H) The abundance of GTP-RhoA in each treatment group, (n=3).