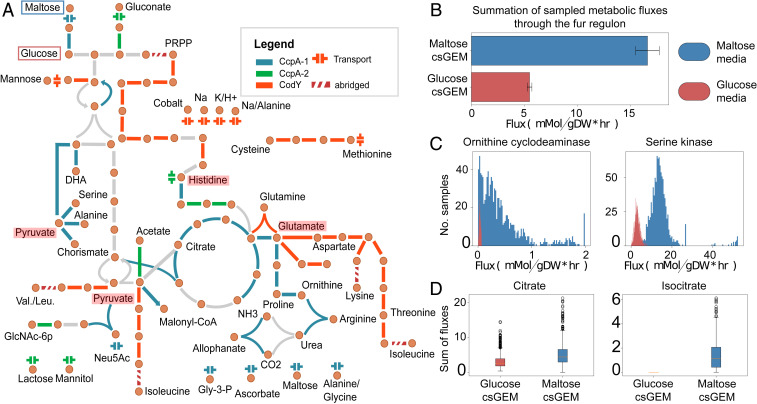
Fig. 3.
Regulation of central metabolism and its interaction with other metabolic subsystems. (A) Overlay of i-modulons onto the map of central metabolism and amino acid metabolism of S. aureus. The two main regulators of these metabolic subsystems, CcpA (blue/green) and CodY (orange), control central carbon and nitrogen metabolism, respectively. These two i-modulons intersect at key metabolic nodes: Pyruvate, histidine, and glutamate (highlighted in red). Entry points of sugars used in the next section, glucose and maltose, are highlighted with red and blue boxes, respectively. (B) Activity of reactions associated with the Fur i-modulon in the presence of different carbon sources, maltose and glucose. The bars represent sum of median sampled fluxes through reactions catalyzed by enzymes in the Fur i-modulon. Unexpected increase in Fur i-modulon activity when carbon source was switched from glucose to maltose is recapitulated through metabolic modeling. (C) Reactions associated with the Fur i-modulon with the largest increase in simulated flux in glucose media. (D) Calculated proxy for intracellular metabolite concentrations.