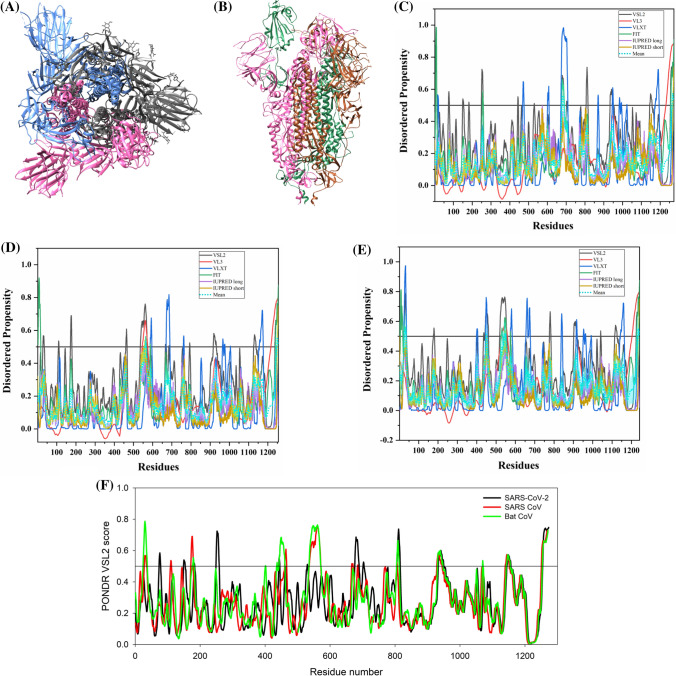
Fig. 3.
Structure and intrinsic disorder propensity of spike glycoprotein (S) from CoVs. a A 3.50 Å resolution structure (PDB ID: 6VSB) of SARS-CoV-2 S obtained through cryo-EM. This homotrimeric structure includes three chains, A (pink), B (dark grey), and C (turquoise). b A 3.6 Å resolution cryo-EM structure (PDB ID: 6ACC) of human SARS S protein complexed with its host-binding partner ACE2. In this structure, three chains are present: A (pink), B (green) and C (dark khaki). Evaluation of intrinsic disorder predisposition in S proteins of c SARS-CoV-2, d human SARS, and e bat CoVs. Graphs c–e depict the disorder profiles generated using six predictors: PONDR® VSL2 (black line), PONDR® VL3 (red line), PONDR® VLXT (blue line), PONDR® FIT (green line), IUPred long (purple line) and IUPred short (golden line). The mean disorder propensity calculated by averaging the disorder scores from all predictors is represented by a short dotted line (sky blue) in graphs. The light sky blue shadow region signifies the mean error distribution. f Aligned disorder profiles generated for all three S proteins is based on the outputs of the PONDR® VSL2