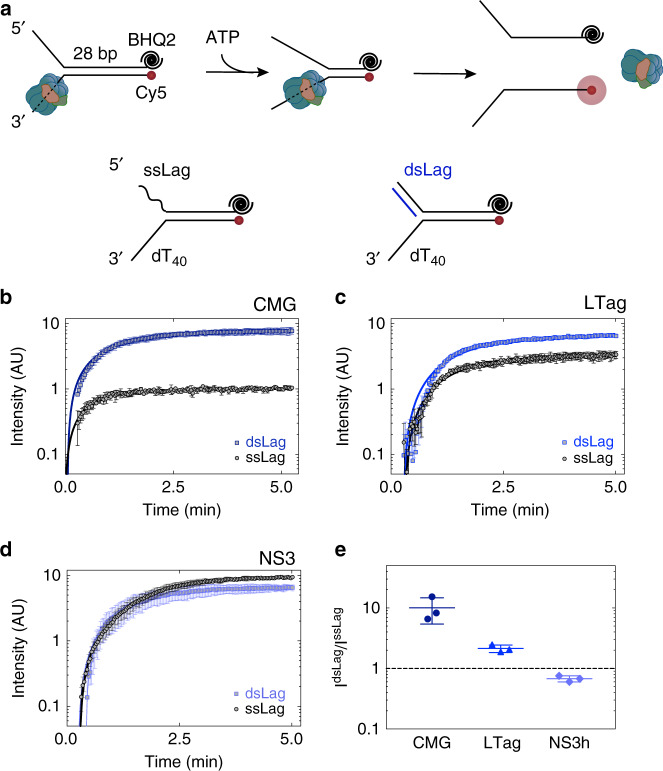
Fig. 3. Duplex DNA engagement by CMG at the fork impedes its helicase activity.
a–d Single-turnover unwinding of fork DNA substrates containing ss (black) or ds (blue) lagging-strand arm by CMG b, large T antigen c, and NS3 d. DNA substrates contained 28-bp duplex region and were modified with Black Hole Quencher 2 (BHQ2) at the 3′-end of the lagging-strand template and Cy5 at the 5′-end of the leading-strand template. Solid lines represent fits to Eq. (2) in b and Eq. (3) in c and d (see Methods). e The ratio of fluorescence plateau intensity on ForkdsLag to ForkssLag indicates that DNA stimulation of fork DNA unwinding by duplexing the lagging-strand arm is helicase dependent. All data represent mean ± SD (n = 3 independent experiments). Source data are provided as a Source Data file.