Fig. 7. CMG bypasses a protein directly crosslinked to the excluded strand with no detectable stalling.
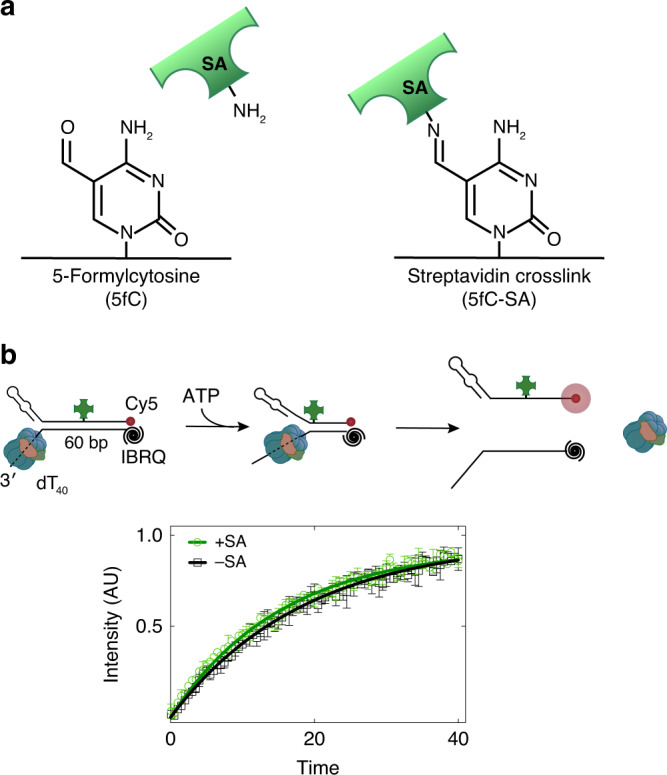
a 5-formyl-cytosine-modified oligonucleotide covalently crosslinks to streptavidin through primary amines. b Single-turnover CMG-mediated unwinding of fork DNA substrates with and without a 5fC-streptavidin cross-link. The 5′ tail of the DNA substrate contained d(GGCA)10 sequence, which generates secondary structures and prevents helicase pausing at the fork junction. Fork DNA included 60-bp duplex region and were modified with Cy5 at the 3′ end of the lagging-strand template and Iowa Black RQ (IBRQ) dark quencher at the 5′ end of the leading-strand template. Separation of the complementary strands by CMG leads to fluorescence increase, a proxy for DNA unwinding. CMG unwinds 5fC-streptavidin-crosslinked and non-crosslinked substrates with the same kinetics. Data represent mean ± SD (n = 3 independent experiments). Solid lines represent fits to Eq. (2) (Methods). Source data are provided as a Source Data file.
