Abstract
Objective
Cross-institutional distributed healthcare/genomic predictive modeling is an emerging technology that fulfills both the need of building a more generalizable model and of protecting patient data by only exchanging the models but not the patient data. In this article, the implementation details are presented for one specific blockchain-based approach, ExplorerChain, from a software development perspective. The healthcare/genomic use cases of myocardial infarction, cancer biomarker, and length of hospitalization after surgery are also described.
Materials and Methods
ExplorerChain’s 3 main technical components, including online machine learning, metadata of transaction, and the Proof-of-Information-Timed (PoINT) algorithm, are introduced in this study. Specifically, the 3 algorithms (ie, core, new network, and new site/data) are described in detail.
Results
ExplorerChain was implemented and the design details of it were illustrated, especially the development configurations in a practical setting. Also, the system architecture and programming languages are introduced. The code was also released in an open source repository available at https://github.com/tsungtingkuo/explorerchain.
Discussion
The designing considerations of semi-trust assumption, data format normalization, and non-determinism was discussed. The limitations of the implementation include fixed-number participating sites, limited join-or-leave capability during initialization, advanced privacy technology yet to be included, and further investigation in ethical, legal, and social implications.
Conclusion
This study can serve as a reference for the researchers who would like to implement and even deploy blockchain technology. Furthermore, the off-the-shelf software can also serve as a cornerstone to accelerate the development and investigation of future healthcare/genomic blockchain studies.
Keywords: blockchain distributed ledger technology, privacy-preserving predictive modeling, online machine learning, clinical information systems, decision support systems
Lay Summary
While multiple healthcare/genomic institutions may want to collaboratively create a predictive model to better predict patient outcomes (eg, heart disease, cancer biomarker, and postsurgery hospital length of stay), privacy concerns of sharing patient data directly and the security risks of having a central server to coordinate the machine learning process can be potential burdens. To mitigate these issues, several cross-institutional distributed predictive modeling methods have been proposed. In this study, the design and implementation details are presented for one specific blockchain-based approach ExplorerChain. The healthcare/genomic use cases for ExplorerChain are also described. Specifically, the 3 main technical components, online learning, blockchain network, and consensus algorithm are introduced. The development configurations, system architecture, and programming languages are described as well. Besides, the designing considerations and the limitations of the implementation were discussed. This study can serve as a reference for researchers who would like to implement/deploy blockchain technology in healthcare/genomic domain. Also, the software of ExplorerChain is available at https://github.com/tsungtingkuo/explorerchain.
BACKGROUND AND SIGNIFICANCE
Cross-institutional predictive modeling is an emerging technology that fulfills both the need for building a more generalizable model and for protecting patient data by only exchanging the models but not the patient data, across multiple healthcare/genomic institutions.1–4 Traditional approaches are mainly based on a client-server architecture, which requires a central server to coordinate the learning process, collect the partially trained models from each site, and then integrate and send the global model back to every site. This centralized approach can create risks such as single-point-of-failure.5,6 Therefore, several existing researches proposed to leverage blockchain,7–9 a peer-to-peer decentralized architecture, to remove the central server.5,6,10–12 Blockchain, a technology originated from financial domain, provides additional desirable technical features such as immutability, provenance, and transparency.11–13
Although the literature illustrated the rationale and results of adopting blockchain for cross-institutional predictive modeling, the details of the implementation are yet to be described. For example, one of the blockchain-based cross-institutional predictive modeling methods, ExplorerChain,14 leverages online machine learning on blockchain and was evaluated on healthcare/genomics datasets (Figure 1). Although the advantages/disadvantages of adopting blockchain, the comparison of different architectures/designs, and the equivalent correctness results of ExplorerChain were shown (more details in Healthcare/genomic use cases section),14 the feasibility study did not include practical considerations while being constructed, and these details could serve as cornerstones for future researchers to develop new algorithms.
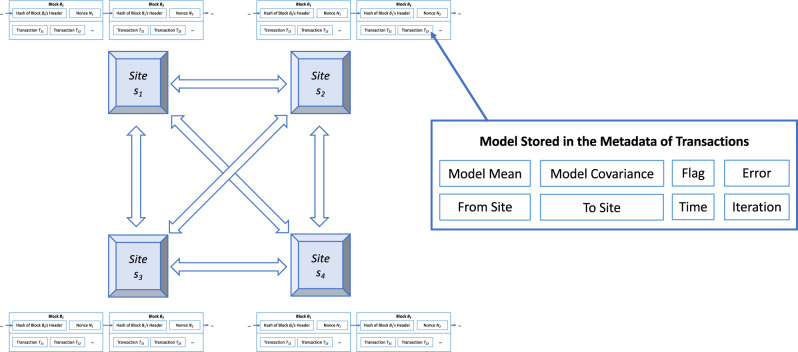
Figure 1.
ExplorerChain.14 Each site maintains an exact copy of the blockchain, and exchanges partially trained machine learning models via the metadata of transactions on-chain. The patient-level data never leave each site to protect patients’ privacy.
OBJECTIVE
In this article, the implementation details for ExplorerChain are dissected from a software development perspective. This article addresses head-on one of the main problems facing blockchain projects today: much is envisioned but little is actually implemented to give developers an idea of using blockchain for distributed predictive model building in practice. A detailed design and implementation of an “off-the-shelf” tool can also help technologists make decisions to adopt blockchain for their specific healthcare/genomic use cases, aiming at accelerating cross-institution research and expedite quality improvement initiatives. ExplorerChain was applied in 3 use cases, including myocardial infarction, cancer biomarker, and length of hospitalization after surgery.14
MATERIALS AND METHODS
The 3 main technical components of ExplorerChain include online machine learning, metadata of transaction, and the Proof-of-Information-Timed (PoINT) algorithm. As shown in Figure 1, online machine learning generates models, metadata of transaction disseminates the models, and the PoINT algorithm determines the learning order. These 3 components are introduced in the following 3 subsections.
Online machine learning and the EXPLORER algorithm
ExplorerChain is designed to operate without a centralized server to compute and manage the predictive models, hence online machine learning4,15–17 is of interest here because it is designed to update the model incrementally and can create/update models without a central server. In ExplorerChain, the EXPLORER online learning method4 was adopted. EXPLORER is an online logistic regression based on a Bayesian approach that can revise the model when the records are updated, without re-training on the entire dataset,4 and therefore is suitable for the purpose of ExplorerChain. The core EXPLORER modeling algorithm, EXPLORER-Intra, was adopted for ExplorerChain, while the inter-site model update component (ie, the central server) of EXPLORER was replaced by transferring the model directly among sites for updates.
Metadata of transaction and blockchain implementation
The partially trained machine learning models are disseminated using the metadata of the blockchain transaction.5,6 The details of the data fields are described in Table 1. Note that in the implementation of this private blockchain network, ExplorerChain only provides sites with nonfinancial incentives (ie, improved correctness of the predictive model using cross-institution data in a privacy-preserving manner), instead of a financial incentive (eg, mining rewards or transaction fees as in Bitcoin), to create the blocks and verify the transactions. MultiChain,18,19 a general purpose blockchain platform, was selected because it is designed for private blockchains and is based on the popular and proven Bitcoin Blockchain platform, according to a prior systematic review of blockchain platforms.9 The default configurations of MultiChain were used to implement ExplorerChain. Also, Mining Diversity,18,19 a round-robin-based consensus protocol in MultiChain designed for private blockchain networks, was adopted.
Table 1.
The on-chain data in ExplorerChain
| Field name | Description | Possible values |
|---|---|---|
| Model mean | The mean vector of the EXPLORER model.4 | A numerical vector with length equal to m+1. |
| Model covariance | The covariance matrix of the EXPLORER model.4 | A numerical (m+1) × (m+1) square symmetric matrix. |
| Flag | The type of action performed by a site. | UNKNOWN, INITIALIZE, UPDATE, EVALUATE, TRANSFER, CONSENSUS, TEST, CLEAR. |
| Error | The training error. | A numerical value between 0 and 1. |
| From site | The site that has submitted the model. | A unique name or identifier representing the sender site. |
| To site | The site that will receive the model. | A unique name or identifier representing the receiver site. |
| Time | The time that the site submitted the model. | A timestamp. |
| Iteration | The current iteration of the cross-institutional model learning process. | A non-negative integer. |
The EXPLORER model contains both the mean coefficient vector and the covariance matrix.4 Also, m is the number of features in the dataset.
PoINT algorithm
The basic idea of the PoINT algorithm is based on the Proof-of-Information (PoI) algorithm5,6: if a site s contains data that cannot be predicted accurately using a current model M, those data may contain more information to improve M; therefore, while choosing the next site to update M, the algorithm should assign s with higher priority. The PoINT algorithm starts with the selection of the best performing model among all sites to prevent the propagation of error. Next, it selects the site with the highest error for the current model, and this site is charged with updating the model. Then, the model update process is repeated, until a site cannot find any other site with higher error; the final consensus model is then complete.
In the original PoI algorithm,5,6 one potential issue is that although the underlying EXPLORER algorithm is guaranteed to converge,4,20 it may require too many iterations (ie, model transferring) without achieving the “best” consensus predictive model, thus consuming unnecessary computational power. To solve this issue, a time-to-leave counter was added to limit the maximum number of iterations in the PoINT algorithm.5,6 This way, ExplorerChain is prevented from executing too long. The training “error” adopted in ExplorerChain was based on the complement of the full Area Under the receiver operating characteristic Curve (AUC).21,22 Specifically, the error was defined as E = 1 – AUCtraining, where AUCtraining was computed using the data at each site. AUC was also used as the evaluation metrics; the training sets were used to guide the process of PoINT (by using AUCtraining), and the test sets were utilized to compute the evaluation metric (ie, AUCtest).
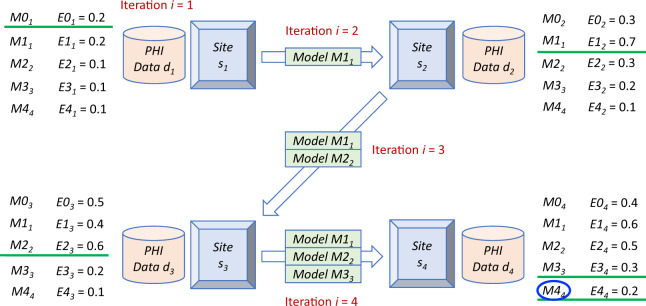
The detailed PoINT algorithm is shown in Algorithms 1–3. Algorithm 1 is the core part of the PoINT algorithm. It determines the machine learning order, and then repeats the training process until finding the consensus model. Each site executes Algorithm 2 (which then executes Algorithm 1) to identify a consensus model for a new blockchain network. A running example for PoINT to find a consensus model is shown in Figure 2. At the initial stage (t = 0), the model with lowest error (site s1 with E01 = 0.2) is selected as the initial model; choosing the best initial model (M01) helps prevent the propagation of error. The selected model M01 is then submitted to site s2, s3, and s4. Next (t = 1), M11 (the same model as M01) is evaluated by each site using the local data. Suppose s2 has the highest error (E12 = 0.7). Given that the data in s2 are less accurately predicted using the model M11, s2 is assumed to contain the richest information to improve M11. Therefore, model M11 is now conceptually “transferred” to s2 within a blockchain transaction (with amount = 0 and transaction fee = 0). Then (t = 2), s2 updates the model as M22. Again, s2 sends M22 to all other sites (within another blockchain transaction), and the site with highest error (or richest information given the current model) will be the next to update the model locally (s3). This process is repeated until a site updates the model and finds itself producing the highest error when compared to other sites, or until the maximum number of iterations is reached. For example, when t = 3, s4 has the highest error (0.3) and thus wins the bid to update the model; but when t = 4, s4 still has the highest error (0.2) using the updated model. Thus, M44 is regarded as the final consensus model and the online machine learning process stops.
Figure 2.
An example of the Proof-of-Information-Timed (PoINT) algorithm.5,6,14Mts is the model and Ets is the error at time t on site s. The timestamp t is not equivalent to the iteration i; the iteration i only increases when the model initialization or transferring occurs. The model/error with green underline is the selected one at that timestamp (ie, at each t, only one model/error is selected). Abbreviation: PHI, Protected Health Information.
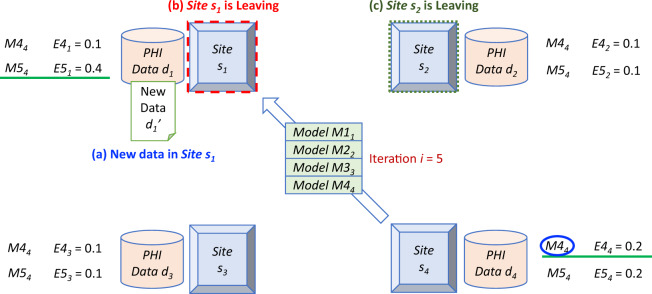
On the other hand, a site executes Algorithm 3 with it has new data (a new site is considered as a site of which all data are new), after a consensus model has been found. Algorithm 3 starts from the latest consensus model and also leverages Algorithm 1 to update the consensus model. The examples for new site/data inclusion and situations in which a site leaves are demonstrated in Figure 3.
Figure 3.
An example of the PoINT algorithm for the situations in which any site adds new data or a site leaves the network.5,6,14 (A) New data (eg, in s1). (B) Site leaving while it is updating the model (eg, s1). (C) Site leaving while not updating the model (eg, s2).
First, if there are new data in s1 (Figure 3A), re-training the whole model is not needed. Instead, an algorithm similar to that shown in Figure 2 can be used to determine whether the model should be updated using the new data. Suppose the new data d1ʹ is in s1, while the current (t = 4) consensus model is M44. In time t = 5, s1 uses the updated data (including both d1 and d1ʹ) to evaluate model M54 (which is the same as M44), and finds that the error E51 = 0.4 is larger than the error of the current updating site (ie, s4 with E54 = 0.2). Therefore, the model M54 is now transferred to s1 to be updated. The iteration i is reset to 1, and the same process shown in Figure 2 is repeated until a final consensus model is identified. In the case that the error E54 is higher than E51, the new data are considered not bringing enough information to update the model M54, thus no transfer/update is required. A similar mechanism can be used for a new site (treated as a site where all data are new).
Next, if the site (eg, s1) leaves while it is updating the model (Figure 3B), it can simply be ignored. This is because until s1 completes the model update, the latest model M54 (at the end of the blockchain) remains unchanged and can be used for prediction tasks by the other sites in the network. Once coming back to the network, s1 can continue the process of updating the model. In the case that the site (eg, s2) leaves while not updating the model (Figure 3C), the departure can be ignored; the site can rejoin the network at any time. For example, if the site with the highest error leaves the network and does not submit its error, it will time out and the site with the second highest error will replace it, and so on. As a result, in both above-mentioned situations, the departure of a site can be dealt with by the blockchain mechanism of ExplorerChain.
There are 4 main hyper-parameters in PoINT: (1) the total number of participating sites N, (2) the polling time period Δ, (3) the waiting time period Θ, and (4) the maximum number of iterations Ω. The total model size (including mean and covariance) is O(m2), where m is the number of features. Using online learning (ie, without a central server like the one in EXPLORER to compute the optimized global model), ExplorerChain is actually an approximation to the optimal solution.
RESULTS
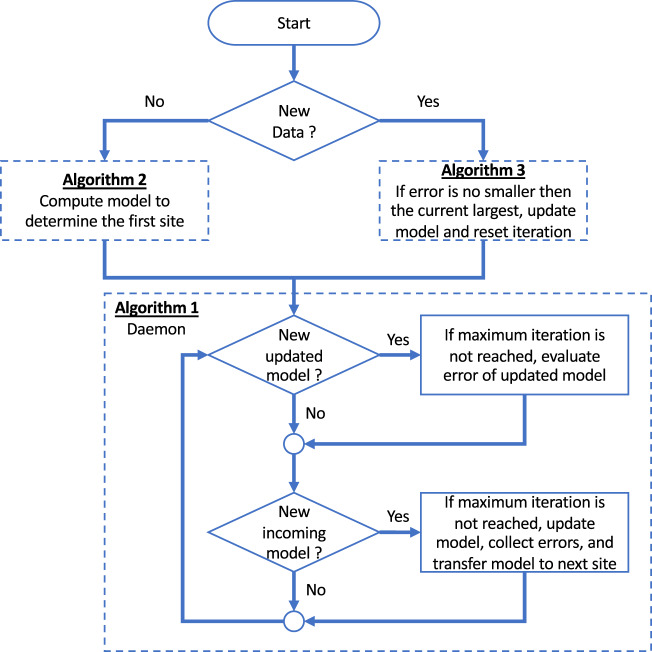
A simplified flowchart of the implemented algorithms is illustrated in Figure 4. When a site starts ExplorerChain, it first determines whether to execute Algorithm 2 (for the initialization scenario) or Algorithm 3 (for the new data scenario). Afterwards, the site runs Algorithm 1 as a “daemon” service to monitor the blockchain network and perform model or error computations as needed. Therefore, Algorithm 1 is always watching the blockchain to check the availability of any newly updated model or an incoming transferred model. In other words, Algorithm 1 keeps running, while occasionally the consensus learning process in it may pause because of the confirmation of a consensus model. Algorithm 1 stops when a site that is running leaves the network (while other sites are still running Algorithm 1), or when the site has new data and would like to stop and run Algorithm 3 instead.
Figure 4.
A simplified flowchart demonstrating the implementation of Algorithms 1–3 of PoINT.
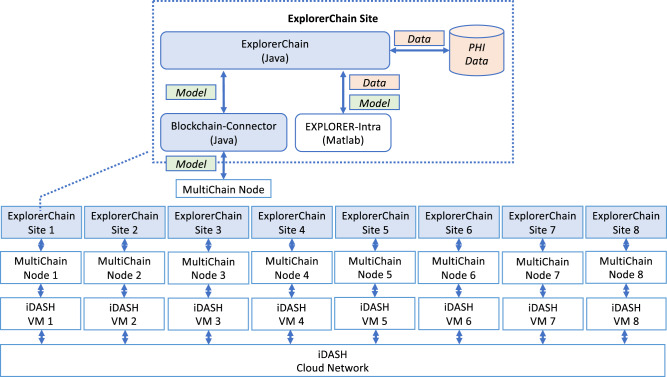
The system architecture and programming languages used in ExplorerChain are demonstrated in Figure 5. The system (including ExplorerChain and the underlying MultiChain) was deployed on integrating Data for Analysis, Anonymization, and SHaring (iDASH),23,24 a private HIPAA-compliant computing environment including both virtual machines (VMs) and a cloud-based network. Within ExplorerChain, Protected Health Information is only used for internal model training (through EXPLORER-Intra), while the model is disseminated on the blockchain (through Blockchain-Connector).
Figure 5.
System architecture of ExplorerChain. This example shows the architecture under the 8-site configuration.
Java is used as the main implementation language to interact with EXPLORER-Intra (written in MATLAB) and MultiChain (written in C++) via runtime command execution. The EXPLORER-Intra MATLAB code was refactored to the APIs (ie, initializing, updating, and evaluating the model) that can be called by ExplorerChain without changing the original learning functionalities. The simulation includes the multiple site scenarios on iDASH VMs, with different numbers of sites (2, 4, and 8 in the experiment). Each VM contains 2 Intel Xeon 2.30 GHz CPUs, 8 GB RAM, and 100 GB storage.
For EXPLORER-Intra, the prior mean was set to 0 and variance was set to 5 for the normal distribution.4 The other hyper-parameters of EXPLORER-Intra were set to the default values of EXPLORER. For ExplorerChain, the hyper-parameter values are as follows: (1) polling time period Δ = 1 (s), (2) waiting time period Θ = 30 (s), (3) maximum iteration Ω = 10, and (4) total number of participating sites N = 2, 4, or 8. The time periods (ie, Δ and Θ) were chosen based on the estimated model training/updating time of EXPLORER-Intra (about 3 s for the experiment datasets in this study, including the time to load MATLAB) and network latency (relatively small in the underlying iDASHcomputing environment). Also, the blockchain network was checked only for the latest N (ie, 2, 4, or 8) transactions that had hexadecimal transaction metadata size >20 in the PoINT algorithm.
HEALTHCARE/GENOMIC USE CASES
The use cases and the efficacy of the ExplorerChain were described in Ref.14 and summarized below (all with 8 total number of participating sites and 80%/20% training/test data splitting for 30 trials): (1) Myocardial infarction.25 In this use case, the binary outcome is the presence of disease, and the dataset contains 9 covariates (eg, Pain in Right Arm) and 1253 samples. ExplorerChain reached a prediction correctness of 0.954 in AUC,21,22 3.633 of average learning iterations, and about 184 s of total execution time.14 (2) Cancer biomarker.26 The binary outcome is the presence of cancer, and the dataset contains 2 covariates (ie, CA-19 and CA-125) and 141 samples. ExplorerChain reached 0.876 in AUC, 3.233 of average learning iterations, and about 173 s of total execution time.14 (3) Length of hospitalization after surgery.27,28 The binary outcome is whether the hospital length of stay is greater than 3 days. This dataset contains 34 covariates (eg, preoperative opioid use) and 960 samples. ExplorerChain reached 0.712 in AUC, 9.833 of average learning iterations, and about 365 s of total execution time.14
DISCUSSION
There are design considerations of ExplorerChain that are common to distributed networks federating data at the institutions of origin: it is based on a semi-trust assumption that the sites are willing to share the aggregated model data but not the patient-level data, and the data format (both syntactic and semantic) on each site must be normalized using standards, such as the Observational Medical Outcomes Partnership (OMOP) Common Data Model (CDM).29 Also, the method is nondeterministic, because the learning process depends on network and computation latency of each blockchain node. Finally, ExplorerChain is platform independent, and can adopt other blockchain such as BigchainDB.30
One potential concern may be the increased model complexity (ie, a large number of covariates). Since ExplorerChain stores model covariance matrix, the space complexity of a model with m covariates is O(m2), as shown in Table 1. In the case of a large m, the size of the model covariance may exceed the limit of a MultiChain. In this case, a different blockchain platform that supports larger transaction size can be adopted.
Regarding limitations, the participants of a permissioned ExplorerChain network are predetermined, therefore the total number of participating sites (the hyper-parameter N) has a known value. Any site within the predetermined participating sites can join or leave the network; however, nonapproved sites cannot join during the process. Besides, with current design, the participating sites cannot join or leave during the initialization phase.
Also, privacy-preserving methods such as the re-identification risks considered in the research field of differential privacy31–37 (ie, the data in some sites are very small thus the model parameters may lead to re-identification of cases and thus compromise privacy) were not fully investigated, while methods such as LearningChain10 focus on protecting the differential privacy. Not only theoretical guarantees of privacy protection, but also ethical, legal, and social implications that may arise from repeated access to a distributed computing system need to be further pondered to protect human subjects.
CONCLUSION
A software implementation of ExplorerChain has been developed, and is publicly available in an open source repository (https://github.com/tsungtingkuo/explorerchain). With the previously shown accuracy, the details about how a blockchain program can be implemented to solve the cross-institutional predictive modeling problem are further described in this study. Also, healthcare/genomic use cases demonstrate the efficacy of ExplorerChain. This work can serve as a reference for the researchers who would like to implement and even deploy blockchain technology, and the off-the-shelf software can also serve as a cornerstone to accelerate the development and investigation of future healthcare/genomic blockchain studies.
FUNDING
T.-T.K. was funded by the U.S. National Institutes of Health (NIH) (OT3OD025462, R00HG009680, R01HL136835, R01GM118609, and U01EB023685) and a UCSD Academic Senate Research Grant (RG084150). The content is solely the responsibility of the author and does not necessarily represent the official views of the NIH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
AUTHOR CONTRIBUTIONS
T.-T.K. contributed in conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, project administration, resources, supervision, software, validation, visualization, and writing (original draft).
ACKNOWLEDGEMENTS
The author would like to thank the Office of the National Coordinator for Health Information Technology (ONC) and the National Institute of Standards and Technology (NIST) for awarding ModelChain, from which ExplorerChain originated, at the 2016 Use of Blockchain in Health IT and Health-related Research Challenge and Workshop. The author would also like to thank Lucila Ohno-Machado, MD, PhD, Rodney A. Gabriel, MD, MAS, Krishna R. Cidambi, MD, Xiaoqian Jiang, PhD, and Shuang Wang, PhD for very helpful discussions, as well as UCSD DBMI and ACTRI teams for the technical support of the iDASH cloud infrastructure.
CONFLICT OF INTEREST STATEMENT
None declared.
REFERENCES
- 1. Jiang W, Li P, Wang S, et al. WebGLORE: a web service for Grid LOgistic REgression. Bioinformatics 2013; 29 (24): 3238–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shi H, Jiang C, Dai W, et al. Secure Multi-pArty Computation Grid LOgistic REgression (SMAC-GLORE). BMC Med Inform Decis Mak 2016; 16 (S3): 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wu Y, Jiang X, Kim J, Ohno-Machado L.. Grid Binary LOgistic REgression (GLORE): building shared models without sharing data. J Am Med Inform Assoc 2012; 19 (5):758–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang S, Jiang X, Wu Y, Cui L, Cheng S, Ohno-Machado L.. Expectation propagation logistic regression (explorer): distributed privacy-preserving online model learning. J Biomed Inform 2013; 46 (3): 480–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kuo T-T, Hsu C-N, Ohno-Machado L. ModelChain: Decentralized Privacy-Preserving Healthcare Predictive Modeling Framework on Private Blockchain Networks. ONC/NIST Use of Blockchain for Healthcare and Research Workshop. September 26–27, 2016. Gaithersburg, MD; 2016.
- 6. Kuo T-T, Ohno-Machado L. ModelChain: decentralized privacy-preserving healthcare predictive modeling framework on private blockchain networks. arXiv preprint: 1802.01746; 2018. https://arxiv.org/abs/1802.01746 Accessed May 15, 2020.
- 7. Nakamoto S. Bitcoin: A Peer-to-Peer Electronic Cash System; 2008. https://bitcoin.org/bitcoin.pdf Accessed May 15, 2020.
- 8. Buterin V. A Next-Generation Smart Contract and Decentralized Application Platform; 2014. https://github.com/ethereum/wiki/wiki/White-Paper Accessed May 15, 2020.
- 9. Kuo T-T, Zavaleta Rojas H, Ohno-Machado L. Comparison of blockchain platforms: a systematic review and healthcare examples. J Am Med Inform Assoc2019; 26 (5): 462–78. [DOI] [PMC free article] [PubMed]
- 10. Chen X, Ji J, Luo C, Liao W, Li P.. When Machine Learning Meets Blockchain: A Decentralized, Privacy-preserving and Secure Design 2018 IEEE International Conference on Big Data (Big Data). December 10–13, 2018. Seattle, WA: IEEE, 2018: 1178–87.
- 11. Kuo T-T, Gabriel RA, Ohno-Machado L. Fair compute loads enabled by blockchain: sharing models by alternating client and server roles. J Am Med Inform Assoc2019; 26 (5): 392–403. [DOI] [PMC free article] [PubMed]
- 12. Kuo T-T, Kim J, Gabriel RA.. Privacy-preserving model learning on blockchain network-of-networks. J Am Med Inform Assoc 2020; 27 (3): 343–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kuo T-T, Kim H-E, Ohno-Machado L.. Blockchain distributed ledger technologies for biomedical and health care applications. J Am Med Inform Assoc 2017; 24 (6): 1211–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kuo T-T, Gabriel RA, Cidambi KR, Ohno-Machado L.. EXpectation Propagation LOgistic REgRession on permissioned blockCHAIN (ExplorerChain): decentralized privacy-preserving online healthcare/genomics predictive model learning. J Am Med Inform Assoc 2020; 27 (5): 747–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yan F, Sundaram S, Vishwanathan S, Qi Y.. Distributed autonomous online learning: regrets and intrinsic privacy-preserving properties. IEEE Trans Knowl Data Eng 2013; 25 (11): 2483–93. [Google Scholar]
- 16. Fontenla-Romero Ó, Guijarro-Berdiñas B, Martinez-Rego D, Pérez-Sánchez B, Peteiro-Barral D.. Online Machine Learning Efficiency and Scalability Methods for Computational Intellect. Hershey, PA: IGI Global; 2013: 27–54. [Google Scholar]
- 17. Shalev-Shwartz S. Online Learning and Online Convex Optimization. FNT in Machine Learning 2011; 4 (2): 107–94. 10.1561/2200000018 [DOI] [Google Scholar]
- 18. Greenspan G. MultiChain Private Blockchain—White Paper; 2015. http://www.multichain.com/download/MultiChain-White-Paper.pdf Accessed May 15, 2020.
- 19.CoinSciencesLtd. MultiChain Open Platform for Blockchain Applications. 2015. http://www.multichain.com Accessed May 15, 2020.
- 20. Minka T. Divergence measures and message passing. Technical Report. Microsoft Research; 2005. https://www.microsoft.com/en-us/research/wp-content/uploads/2016/02/tr-2005-173.pdf Accessed May 15, 2020.
- 21. Lasko TA, Bhagwat JG, Zou KH, Ohno-Machado L.. The use of receiver operating characteristic curves in biomedical informatics. J Biomed Inform 2005; 38 (5): 404–15. [DOI] [PubMed] [Google Scholar]
- 22. Hanley JA, McNeil BJ.. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 1982; 143 (1): 29–36. [DOI] [PubMed] [Google Scholar]
- 23. Ohno-Machado L, Bafna V, Boxwala A, et al. iDASH. Integrating data for analysis, anonymization, and sharing. J Am Med Inform Assoc 2012; 19: 196–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ohno-Machado L. To share or not to share: that is not the question. Sci Transl Med 2012; 4 (165): 165cm15. [DOI] [PubMed] [Google Scholar]
- 25. Kennedy R, Fraser H, McStay L, Harrison R.. Early diagnosis of acute myocardial infarction using clinical and electrocardiographic data at presentation: derivation and evaluation of logistic regression models. Eur Heart J 1996; 17 (8): 1181–91. [DOI] [PubMed] [Google Scholar]
- 26. Zou KH, Liu A, Bandos AI, Ohno-Machado L, Rockette HE.. Statistical Evaluation of Diagnostic Performance: Topics in ROC Analysis. Boca Raton, FL: CRC Press; 2011. [Google Scholar]
- 27. Sharma BS, Swisher MW, Doan CN, Khatibi B, Gabriel RA.. Predicting patients requiring discharge to post-acute care facilities following primary total hip replacement: does anesthesia type play a role? J Clin Anesth 2018; 51: 32–6. [DOI] [PubMed] [Google Scholar]
- 28. Gabriel RA, Waterman RS, Kim J, Ohno-Machado L.. A predictive model for extended postanesthesia care unit length of stay in outpatient surgeries. Anesth Analg 2017; 124 (5): 1529–36. [DOI] [PubMed] [Google Scholar]
- 29.TheOHDSICommunity. The Book of OHDSI: Observational Health Data Sciences and Informatics; 2020. https://ohdsi.github.io/TheBookOfOhdsi/TheBookOfOhdsi.pdf Accessed May 15, 2020.
- 30. McConaghy T, Marques R, Müller A, et al. BigchainDB: A Scalable Blockchain Database; 2016. https://www.bigchaindb.com/whitepaper/ Accessed May 15, 2020.
- 31. Dwork C. Differential privacy: A survey of results. International conference on theory and applications of models of computation. Berlin: Springer; 2008: 1–19. [Google Scholar]
- 32. Dwork C, Roth A.. The Algorithmic Foundations of Differential Privacy. FNT in Theoretical Computer Science 2013; 9 (3-4): 211–407. 10.1561/0400000042 [DOI] [Google Scholar]
- 33. Ji Z, Jiang X, Wang S, Xiong L, Ohno-Machado L.. Differentially private distributed logistic regression using private and public data. BMC Med Genomics 2014; 7 (Suppl 1): S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jiang X, Ji Z, Wang S, Mohammed N, Cheng S, Ohno-Machado L.. Differential-private data publishing through component analysis. Trans Data Priv 2013; 6 (1): 19. [PMC free article] [PubMed] [Google Scholar]
- 35. Li H, Xiong L, Jiang X.. Differentially Private Histogram and Synthetic Data Publication. Medical Data Privacy Handbook. Cham: Springer; 2015: 35–58. [Google Scholar]
- 36. Li H, Xiong L, Jiang X, Liu J. Differentially private histogram publication for dynamic datasets: an adaptive sampling approach. Proceedings of the 24th ACM International on Conference on Information and Knowledge Management: ACM, 2015: 1001–10. [DOI] [PMC free article] [PubMed]
- 37.McSherry F, Talwar K. Mechanism Design via Differential Privacy. 48th Annual IEEE Symposium on Foundations of Computer Science (FOCS'07): IEEE, 2007: 94–103. [Google Scholar]