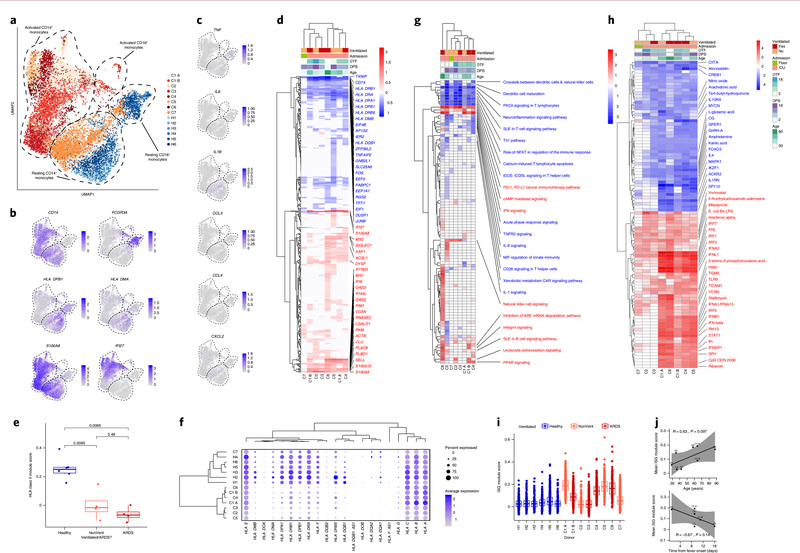
Fig. 2 |. Robust HLA class II downregulation and type I interferon-driven inflammatory signatures in monocytes are characteristics of SARS-CoV-2 infection.
a, UMAP embedding of all monocytes colored by sample of origin. n = 10,339 cells are plotted from n = 14 biologically independent samples. b, UMAP embedding of monocytes colored by CD14 and FCGR3A (encoding CD16a, to distinguish between CD14+ and CD16+ monocytes), HLA-DPB1 and HLA-DMA (illustrating HLA class II downregulation in patients with COVID-19) and S100A9 and IFI27 (demonstrating canonical inflammatory signatures in patients with COVID-19). c, UMAP embedding of monocytes colored by genes encoding pro-inflammatory cytokines previously reported to be produced by circulating monocytes in severe COVID-196, namely TNF, IL6, IL1B, CCL3, CCL4 and CXCL2. d,g,h, Heatmaps of DE genes (d), differentially regulated canonical pathways (g) and differentially regulated predicted upstream regulators (h) between CD14+ monocytes of each COVID-19 sample compared to CD14+ monocytes of all healthy controls. The heatmap in d is colored by average log(fold-change), while heatmaps in g and h are colored by z-score. All displayed genes, pathways and regulators are statistically significant at the P < 0.05 confidence level by Seurat’s implementation of the Wilcoxon rank-sum test (two-sided, adjusted for multiple comparisons using Bonferroni’s correction, in d) or Ingenuity Pathway Analysis (IPA) implementation of the Fisher exact test (right-tailed, in g and h). The 50 genes (d), 25 pathways (g) or 50 regulators (h) with the highest absolute average log(fold-change) or z-score across all donors are labeled. Genes with a net positive average log(fold-change) or z-score are labeled in red; genes with a net negative average log(fold-change) or z-score are labeled in blue. DPS, days post-symptom onset; DTF, days from first reported or measured fever. e, Boxplot showing the mean HLA class II module score of CD14+ monocytes from each sample, colored by healthy donors (blue), non-ventilated patients with COVID-19 (orange) or ventilated patients with COVID-19 (red). Shown are exact P values by two-sided Wilcoxon rank-sum test. n = 6, n = 4 and n = 4 biologically independent samples for Healthy, NonVent and ARDS, respectively. f, Dot plot depicting percent expression and average expression of all detected HLA genes in CD14+ monocytes by donor. i, Boxplot showing the IFNA module score of each cell, colored by healthy donors (blue), non-ventilated patients with COVID-19 (orange) or ventilated patients with COVID-19 (red). j, Scatter plots depicting the correlation between the mean ISG module score of CD14+ monocytes in each sample and the patient age (top) and time–distance from first measured or reported fever (bottom). Shown are Pearson’s r, exact two-sided P values and the 95% confidence interval. n = 8 (top) and n = 6 (bottom) independent biological samples. Number of cells for d,f–i: C1 A, 1,561; C1 B, 1,858; C2, 217; C3, 1,102; C4, 713; C5, 462; C6, 277; C7, 2,095; H1, 680; H2, 325; H3, 215; H4, 166; H5, 444; H6, 224. For d,g–h, cells from all healthy controls (n = 2,054 cells) are used to generate comparisons with each COVID-19 sample. For e,i, boxplot features: minimum whisker, 25th percentile − 1.5 × IQR or the lowest value within; minimum box, 25th percentile; center, median; maximum box, 75th percentile; maximum whisker, 75th percentile + 1.5 × IQR or greatest value within.