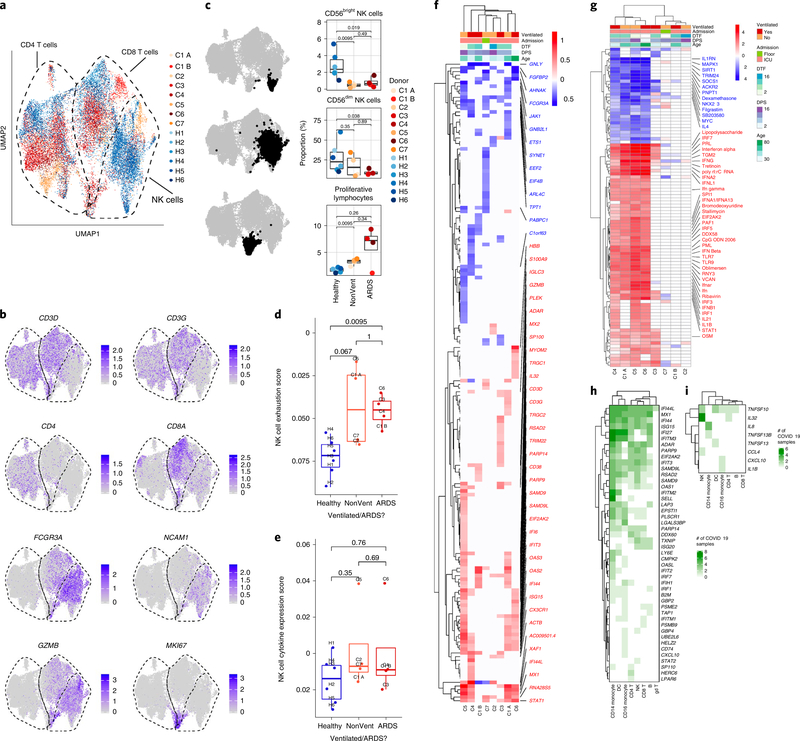
Fig. 3 |. Heterogeneous patterns of NK cell exhaustion and IFN response in COVID-19.
a, UMAP embedding of CD4+ T cells, CD8+ T cells and NK cells colored by sample of origin. b, UMAP embedding colored by lineage genes (CD3D, CD3G, CD4, CD8A, FCGR3A and NCAM1) and selected functional/phenotypic markers (GZMB and MKI67). For a,b, n = 22,016 cells are plotted from n = 14 biologically independent samples. c, Boxplots depicting proportions of CD56dim NK cells, CD56bright NK cells and proliferating lymphocytes among total T and NK cells by sample of origin. The cells used to calculate each proportion are highlighted in bold black in the adjacent UMAP embeddings and were identified by manually labeling clusters generated by clustering CD4+ T cells, CD8+ T cells and NK cells alone. Shown are exact two-sided P values from the Wilcoxon rank-sum test. n = 386 (top), n = 4,899 (middle), n = 781 (bottom) total from n = 6, n = 4 and n = 4 biologically independent samples for Healthy, NonVent and ARDS, respectively. d, Boxplot showing the mean expression score by only NK cells of three canonical markers of NK cell exhaustion: LAG3, PDCD1 (encoding PD-1) and HAVCR2 (encoding TIM-3). Shown are exact two-sided P values by Wilcoxon rank-sum test. e, Boxplot showing the mean expression score by only NK cells of four canonical NK cell cytokine genes (CCL3, CCL4, IFNG and TNF). Shown are exact P values by Wilcoxon rank-sum test. For d,e, n = 6, n = 4 and n = 4 biologically independent samples for Healthy, NonVent and ARDS, respectively. In c–e, boxplot features: minimum whisker, 25th percentile − 1.5 × IQR or the lowest value within; minimum box, 25th percentile; center, median; maximum box, 75th percentile; maximum whisker, 75th percentile + 1.5 × IQR or greatest value within. f,g, Heatmaps of DE genes (f) and differentially regulated predicted upstream regulators (g) between NK cells of each COVID-19 sample compared to NK cells of all healthy controls. As in Fig. 2, f is colored by average log(fold-change), while g is colored by z-score. All displayed genes and regulators are statistically significant at the P < 0.05 confidence level by Seurat’s implementation of the Wilcoxon rank-sum test (two-sided, adjusted for multiple comparisons using Bonferroni’s correction, f) or IPA’s implementation of Fisher exact test (right-tailed, g). The 50 genes or regulators with the highest absolute average log(fold-change) or z-score across all donors are labeled. Genes with a net positive average log(fold-change) or z-score are labeled in red; genes with a net negative average log(fold-change) or z-score are labeled in blue. DPS, days post-symptom onset; DTF, days from first reported or measured fever. Number of cells for f,g: C1 A, 354; C1 B, 387; C2, 271; C3, 328; C4, 104; C5, 518; C6, 58; C7, 130; cells from all healthy controls (n = 4,707 cells) were used to generate comparisons with each COVID-19 sample. h,i, Heatmaps of differentially upregulated ISGs (h; Supplementary Table 25) and cytokines (i; Supplementary Table 25) in donors with COVID-19, colored by the number of COVID-19 samples in which the gene was differentially expressed relative to all healthy controls. DE genes used to construct these heatmaps are provided in Supplementary Tables 4–10. An ISG or cytokine was counted as differentially expressed if it had an average log(fold-change) > 0.25 and an adjusted two-sided P value < 0.05 by Seurat’s implementation of the Wilcoxon rank-sum test. n = 8 biologically independent COVID-19 samples compared to n = 6 biologically independent healthy controls.