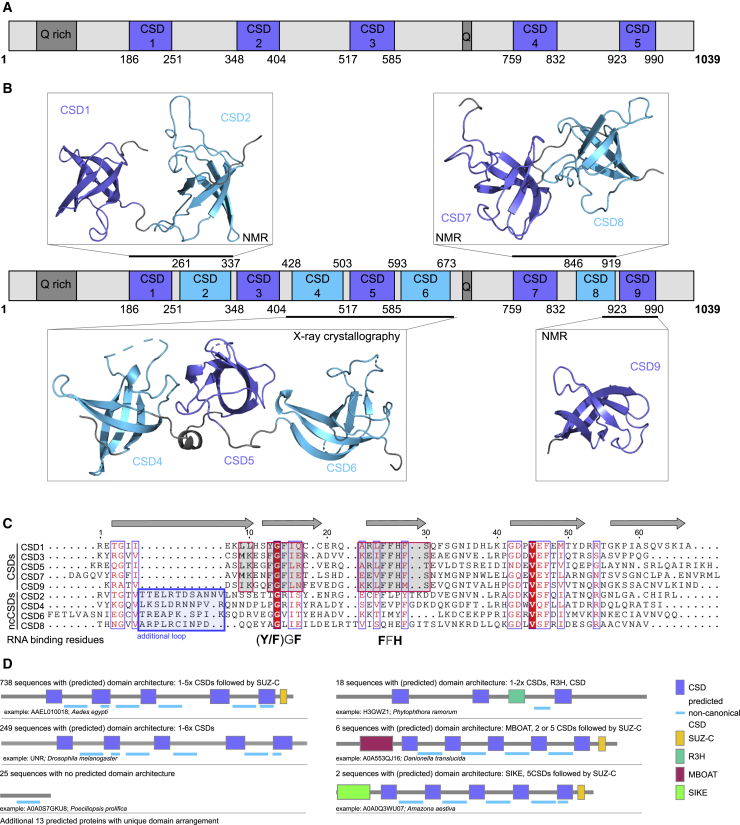
Figure 1.
Revised Domain Assignment in Drosophila Unr (dUnr)
(A) Hitherto domain arrangement scheme of Drosophila Unr that shows the distribution of the five canonical CSDs that have been previously annotated (1, 2, 3, 4, and 5).
(B) Revised domain arrangement scheme (middle) that shows the distribution of previous CSDs (now numbered 1, 3, 5, 7, and 9; purple) and the four non-canonical CSDs (ncCSDs) we discovered in this work (2, 4, 6, and 8; cyan). (Clockwise) NMR solution structures of dUnrCSD12 (aa 179–344; PDB: 6Y6M), CSD78 (aa 756–922; PDB: 6Y4H), CSD9 (aa 899–989; PDB: 6Y96), and a crystal structure of CSD456 (aa 424–677; PDB: 6Y6E), all determined in this study, are arranged around the revised domain arrangement scheme; note that only a single representative NMR assemble structure is shown per construct for clarity, and ensembles are provided in Figure S1.
(C) Sequence alignment of Drosophila canonical CSDs and ncCSDs. The same or similar residues between all domains are colored; similar residue regions between the canonical CSDs are highlighted by red boxes, which align with the two RNA-binding regions (Y/FGF and FFHF). Additional loops only present in ncCSDs are highlighted by a blue box. The alignment has been done using Emboss Needle (Madeira et al., 2019), and ESPript (Robert and Gouet, 2014) has been used for illustration.
(D) Search results of UniProtKB, using a new hidden Markov model based on an ncCSD sequence alignment (Figure S1J). Examples of different protein families containing the predicted ncCSDs are highlighted.