Figure 2.
Co-crystal Structure of the DBD of Caulobacter ParB with parS
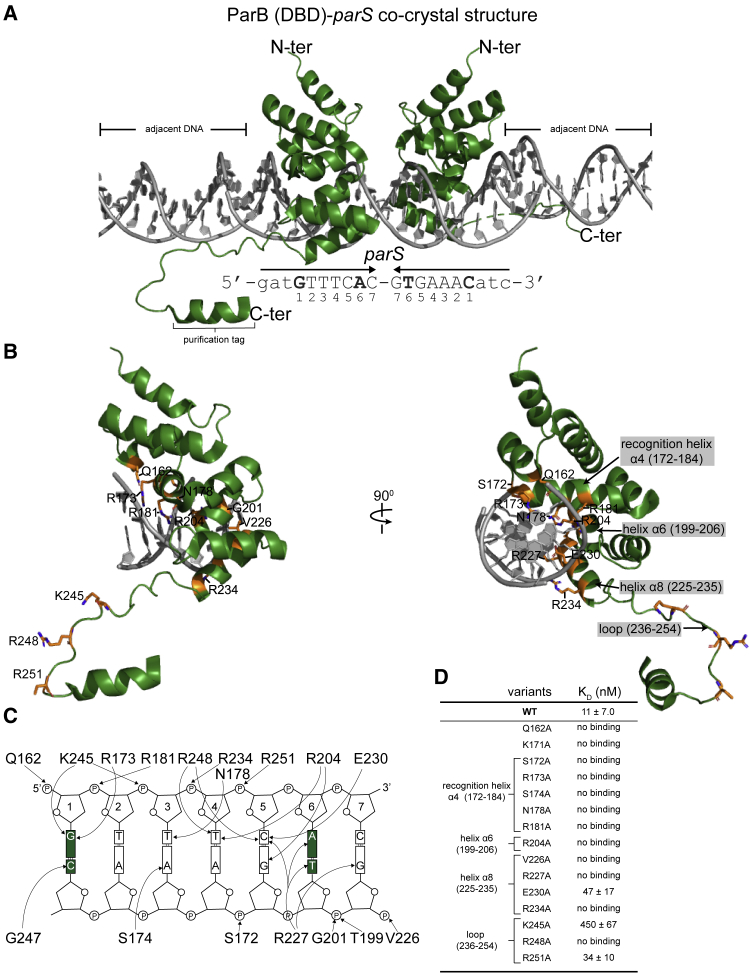
(A) The 2.4-Å resolution structure of two ParB (DBD) monomers (dark green) in complex with a 20-bp parS DNA (gray). The nucleotide sequence of the 20-bp parS is shown below the structure; bases (guanine 1 and adenine 6) that are different from NBS are in bold. The purification tag is also visible in one of the DBD monomers. Loop (236–254) contacts the adjacent DNA in the crystal lattice.
(B) One monomer of ParB (DBD) is shown in complex with a parS half-site; residues that contact the DNA are labeled and colored in orange.
(C) Schematic representation of ParB (DBD)-parS interactions. For simplicity, only a parS half-site is shown. The two bases at position 1 and 6 that are different between parS and NBS are highlighted in dark green.
(D) Alanine scanning mutagenesis and the in vitro dissociation constant (KD) ± standard deviation (SD) of ParB variants to parS DNA. See also STAR Methods for details on curve fitting and calculation of SD values.